15 Pollution and Bioremediation
Pollution and Bioremediation
Achieving sustainable life, for humans, animals, and the environment, requires a plan of action to mitigate anthropogenically-induced damage and develop future practices to maintain planetary homeostasis. One of the biggest threats to ourselves and the environment is the buildup of human waste and pollution (e.g. plastics, oil, synthetic products, etc.) that take a tremendously long time to naturally breakdown. With the human population predicted to continually climb, continuing the same destructive practices will only result in more waste generation in a fraction of the time that it takes for decomposition. One alternative means to solve the pollution problem is to use specialized microbiomes for bioremediation. That is, a specially designed microbial consortia could clean up (i.e. breakdown and assimilate) various toxic molecules much more quickly. However, this will take time to understand how and which type of microbes could perform these tasks and if there are any associated (negative) effects.
Alternative Strategies for Microbial Remediation of Pollutants via Synthetic Biology
Article by Jaiswal and Shukla, 2020 licensed under the terms of the Creative Commons Attribution License (CC BY).
Continuous contamination of the environment with xenobiotics and related recalcitrant compounds has emerged as a serious pollution threat. Bioremediation is the key to eliminating persistent contaminants from the environment. Traditional bioremediation processes show limitations, therefore it is necessary to discover new bioremediation technologies for better results. In this review we provide an outlook of alternative strategies for bioremediation via synthetic biology, including exploring the prerequisites for analysis of research data for developing synthetic biological models of microbial bioremediation. Moreover, cell coordination in synthetic microbial community, cell signaling, and quorum sensing as engineered for enhanced bioremediation strategies are described, along with promising gene editing tools for obtaining the host with target gene sequences responsible for the degradation of recalcitrant compounds. The synthetic genetic circuit and two-component regulatory system (TCRS)-based microbial biosensors for detection and bioremediation are also briefly explained. These developments are expected to increase the efficiency of bioremediation strategies for best results.
Introduction
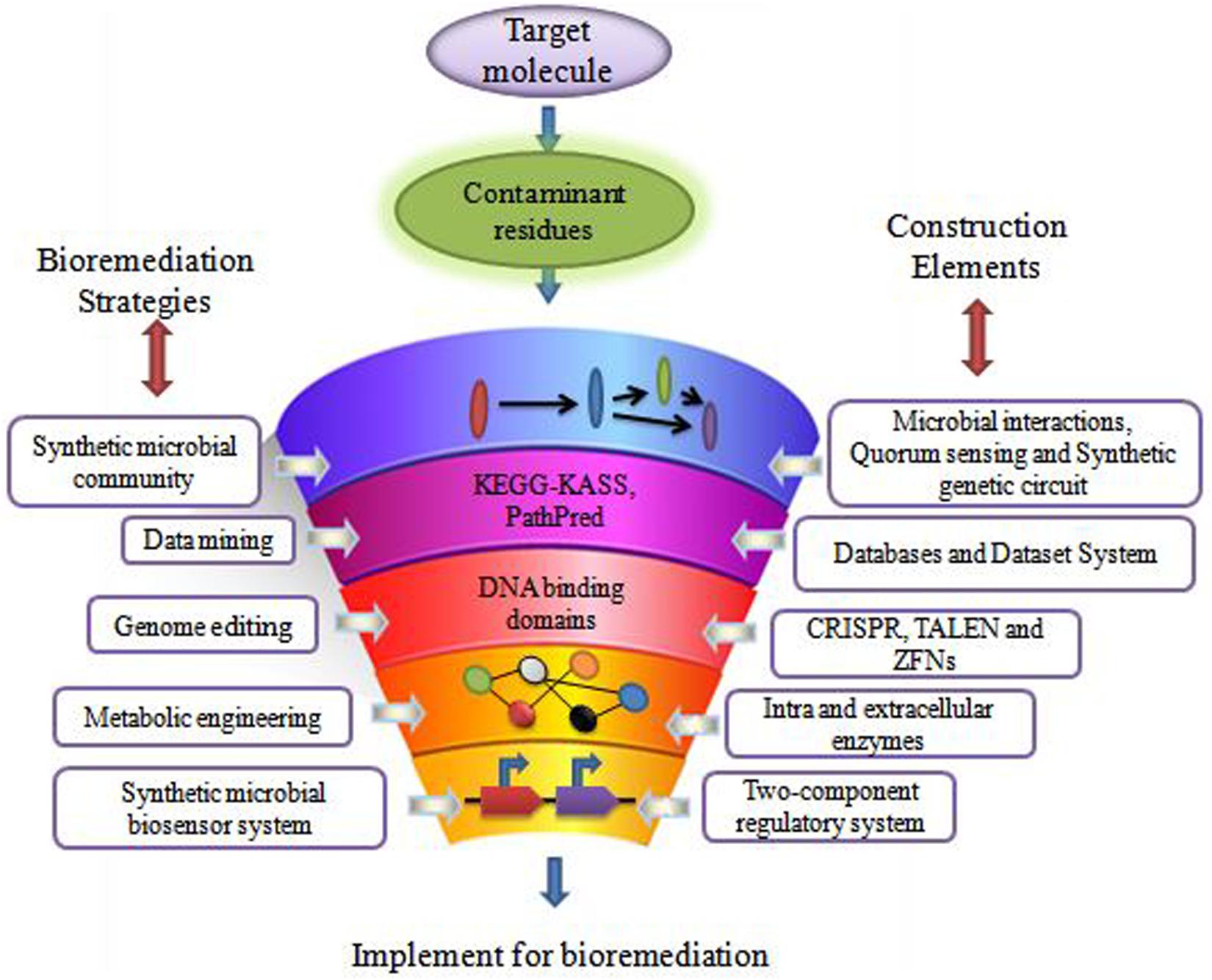
The remediation processes aided by microorganisms present at the various contaminated scenarios constitute bioremediation (Basu et al., 2018; Kumar et al., 2019). Microbial remediation uses multiple metabolic pathways responsible for enzyme production (Sharma B. et al., 2018; Dangi et al., 2019). These enzymes mainly take part in the degradation pathways of xenobiotics (Junghare et al., 2019). There are different customary methods for bioremediation, primarily based on the site of bioremediation, in and ex situ (Tomei and Daugulis, 2013). In situ is applied to the site to minimize soil disturbance. This method is mostly adopted due to less expenditure from avoiding excavation and transport of contaminated soil (Khan et al., 2004). According to Khan et al., 2004 less disruption in in situ bioremediation causes less dust dispersion and hence better degradation (Joshi et al., 2016) of contaminant. Bioaugmentation, bioventing, biosparging, and engineered in situ bioremediation are main in situ bioremediation methods (Azubuike et al., 2016). Ex situ bioremediation methods are solid phase system (composting, landfarming, and biopiling) and slurry phase system (bioreactors) (Kumar et al., 2011). Transportation of soil to accelerate microbial degradation are done by solid and slurry phase systems, whereby treatments of domestic, industrial, and organic waste are done by ex situ bioremediation (Juwarkar et al., 2010). These traditional bioremediation methods take time and consume much cost expenditure, giving less result output. Traditional bioremediation (Duarte et al., 2017) processes showed the above limitations of extra time taking, less removal or dissimilation of pollutants, (Bharagava et al., 2019) disturbance to nature delicacy such as more land coverage for a long time, and a foul smell in the environment (Dangi et al., 2019; Kumar, 2019). Therefore, researchers are eager to discover new bioremediation technologies for best results. Dvoøák et al. (2017) described bioremediation via synthetic biology for boosting bioremediation strategies. This approach can catch the catabolic (Jacquiod et al., 2014) and metabolic complexities for reviewing the potential of the microbial population synthetically. The preliminary information for developing synthetic microbial models for bioremediation can be obtained by mining genes from the databases (Fajardo et al., 2019). The computer logics involvement can determine the microbial cell interactions with recalcitrant compounds (Kim et al., 2015). These strategies can together grasp the natural metabolic potential of microorganisms to transform into novel biological entities of interest (Dangi et al., 2019). Furthermore, the regulation of metabolic pathways (Alves et al., 2018) in a controlled manner can also be achieved for bioremediation processes (Rochfort, 2005). This transition via synthetic biology application (Figure 1) for remediation purposes would improve the bioremediation processes via the involvement of potent (Zhu et al., 2017) dissimilating particular contaminants (Trigo et al., 2008). Synthetic biological systems mediate cellular modulations for efficient functioning and working of existing processes. They permit the modification of cellular processes viz. metabolic pathway acting for a particular chemical compound. The advancement of synthetic biology for bioremediation of various contaminants is attaining the focus of scientists and researchers. For instance, a sustainable synthetic microbial community’s establishment for bioremediation is being investigated. Microbial interactions and quorum sensing within communities are vastly studied for application in the area of bioremediation with synthetic biology applications. Achievement of the synthetic genetic circuit of Pseudomonas putida proved to be the golden gadget for degradation studies. Besides this, genome editing by CRISPR-Cas, TALEN, and ZFNs adds knowledge for reviewing the progression in bioremediation studies. Synthetic microbial biosensors and metabolic engineering of cellular processes for utilization and detection of contaminant residues will remediate the environment from persistent recalcitrant pollutants. This review is focused on the above mentioned strategies and their elements (Figure 2) applicable for bioremediation purposes and research.
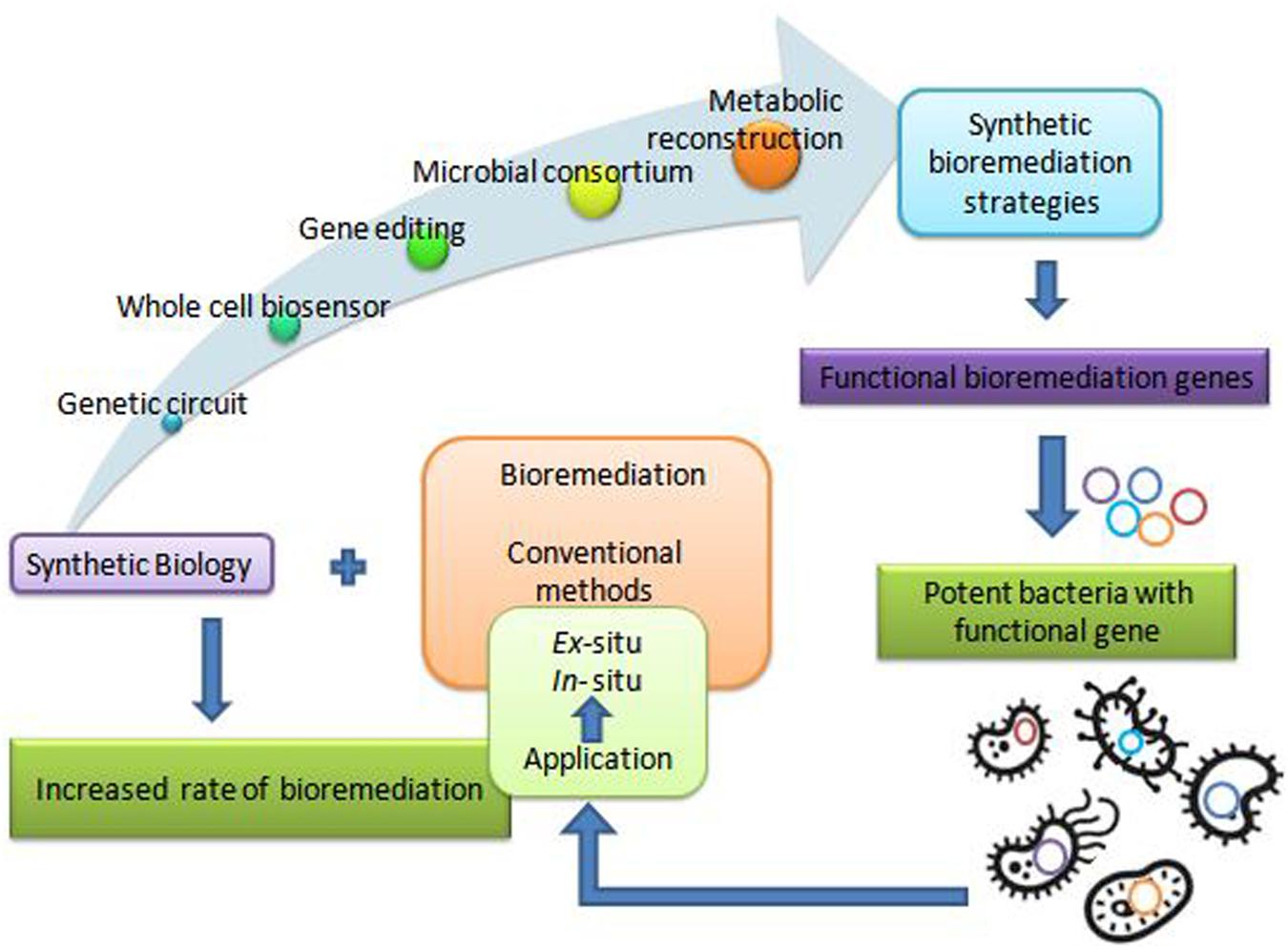
Metabolic Reconstruction for Designing Synthetic Models
A computational platform is utilized for the reconstruction of cellular metabolism (Agapakis et al., 2012) via metabolic pathway analysis (MPA) (Banerjee et al., 2016). MPA mathematically represents the reactions of metabolism. This method is based on stoichiometric balance reactions so as to propose steady-state metabolic flux during cellular growth. The stoichiometry matrix imposes constraints of flux, making the consumption and production of the compound at a steady state (Bordbar et al., 2014). The maximum and minimum flux of a reaction can also be determined by providing the topper and least bound. This helps to define the extent of permissible flux supply (Richelle et al., 2016; Rawls et al., 2019). The next step is defining the objective according to the biological problem to be studied. This objective mathematically represents the reactions responsible for the phenotype appearance. The mathematical reactions and phenotype are combined with linear equations and solved by computational algorithms such as COBRA Toolbox11 and Matlab toolbox (Orth et al., 2010; Bordel, 2014). FBA is fundamentally simple, having immense applications in studying gaps, physiology, and genomes via systems biology approach (Kim et al., 2015; Hellweger et al., 2016). These gaps are missing metabolic reactions, making the genome partially known. FBA uses computational algorithms that can predict missing reactions viz. OptKnock and OptCom, which can knock out the genes responsible for producing the desired compound (Biggs et al., 2015). These approaches are beneficial for constructing microbial communities for bioremediation of particular contaminants (Khandelwal et al., 2013). However, MPA is the most challenging method when metabolic information is incomplete, making it difficult to obtain a real model (Covert et al., 2001). But this method shows cellular functions in the dynamic community, and thus is very useful for the prediction and exchange of metabolic flux in communities of microorganisms (Khandelwal et al., 2013). Recently, a metabolic model has been constructed by using two Geobacter species with parameterized electron transfer and metabolic exchange to characterize syntrophic growth dynamics. Such a system may have useful applications in the field of bioremediation and degradation of particular contaminants (Butler et al., 2010). A computational platform is also needed for better prediction of engineered genetic pathways for community dynamics. A graph-based tool Metabolic Tinker was developed by McClymont and Soyer to identify thermodynamically feasible biochemical routes for compounds deterioration (Johns et al., 2016). This may be applied to identify the routes for degradation of recalcitrant compounds by microbial consortia. These computational tools are utilized along with omics (Kim et al., 2014; El Amrani et al., 2015) and biological data for desired output (Berger et al., 2013) and toxicity prediction (i.e., META-CASETOX System) (Peijnenburg and Damborský, 2012). These are also applied for functional gene identification and their profile analysis, PCR analysis and drug discovery, etc (Dangi et al., 2019). Computer-aided drug discovery and development (CADDD) is used effectively with chemical and biological aspects, i.e., chemical structures accounting the biological role and its activity via ligand-based drug design, structure-based drug design, quantitative structure-property relationships, and quantitative structure-activity (Kapetanovic, 2008). Furthermore, Table 1 depicts similar methodologies applicable to bioremediation studies. De Jong (2002) analyzed the multicellular feedback control strategy in a bacterial consortium (Bruneel et al., 2011) to define the robustness conceivable under desired conditions.
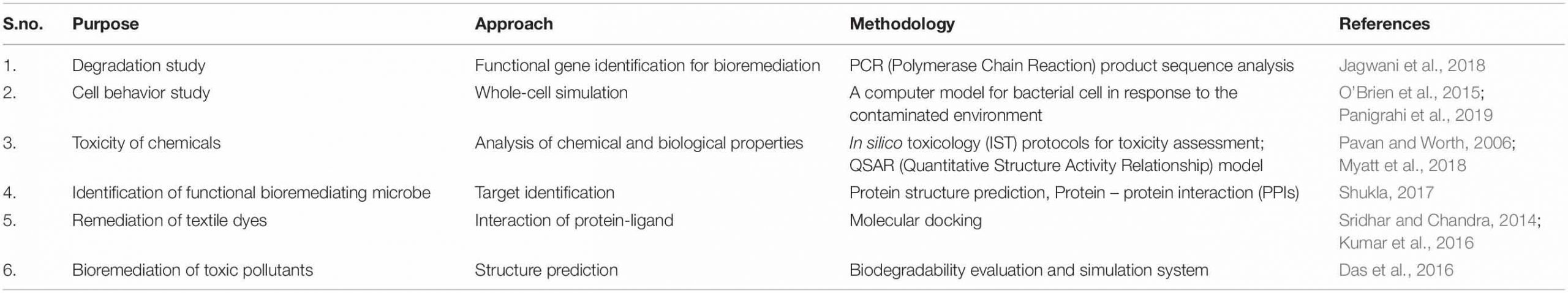
They utilized an ordinary differential equations (ODE)-based model and agent-based simulation on a consortium (Hawley et al., 2014; Atashgahi et al., 2018) of interacting species population for increasing the efficacy of the proposed feedback control strategy. The application of bioinformatics (Arora and Bae, 2014) resources is a prerequisite dimension for obtaining the data to begin the microbial bioremediation studies of recalcitrant compounds (Gong et al., 2012; Ofaim et al., 2019). This involves the information related to the degradation of xenobiotics by microbes and their pathways for dissimilation (Dao et al., 2019; Salam and Ishaq, 2019; Thelusmond et al., 2019; Wei et al., 2019). The data related to end products and intermediate metabolites released throughout degradation pathways can also be retrieved (Dvoøák et al., 2017). An extended information source linked to degradation is MetaRouter, allowing data (Singh and Gothalwal, 2018) for life sciences laboratories to explore degradation possibilities of recalcitrant compounds (Mohanta et al., 2015). The information on oxygenic degradation of xenobiotics can be retrieved from OxDBase, a biodegradative oxygenase database (Chakraborty et al., 2014; Shah et al., 2018). Oxygenase is a class of enzyme which transfers the oxygen molecule for oxidizing the chemical compound. They play a role in the degradation of organic compounds by aromatic ring cleavage (Jadeja et al., 2014). OxDBase is very particular in providing knowledge of oxygenases-catalyzed reactions, and is a powerful tool applicable to bioremediation studies (Singh, 2018). Bioconversion and biodegradation of persistent and toxic xenobiotics (Desai et al., 2010; Bao et al., 2017) compounds catalyzed by oxygenases decrease the compound sustainability and toxicity in the environment (Kües, 2015; Kondo, 2017). Therefore, OxDBase is very helpful in acknowledging the degradation processes involved in bioremediation (Shah et al., 2012). The transcriptional characterization of genes responsible for the biodegradation and biodissimilation of a particular compound has great significance in proposing molecular methodologies. This can be done by the Bionemo (Biodegradation Network Molecular Biology) database (Libis et al., 2016a). Bionemo contains the entries for sequences of genes coding for biodegradation (Carbajosa and Cases, 2010). It also links the gene transcription and its regulation (Libis et al., 2016b). The data retrieved from Bionemo can be used for designing cloning experiments and primers (Arora and Shi, 2010). Garg et al. (2014) used eMolecules and the EAWAG-BBD PPS database for the prediction of pathways involved in the biodegradation of 1-naphthyl-N-methyl carbamate. These above findings empower the researchers to analyze and establish what prerequisites must be fulfilled for developing synthetic bioremediation models.
Designing the Synthetic Microbial Communities
Recent advancements in the field of synthetic biology for environmental issues have shown a great impact. The use of GMOs in environmental biotechnology for remediation (Malla et al., 2018) of toxic compounds, xenobiotics, and pesticidal compounds are being done. To design a synthetic community, it is important to understand natural microbial communities (Schloss and Handelsman, 2008). In a natural community, it is difficult to find out which species are actually taking part in bioremediation (Großkopf and Soyer, 2014). Thus, a synthetic microbial community is a promising method for constructing an artificial microbial community with function-specific species for bioremediation purposes. These communities may act as a model system for the study of functional, ecological, and structural characteristics in a controlled manner. Großkopf and Soyer (2014), defined synthetic community by the culturing of two microbial species under well-defined conditions on the basis of interaction and function (Bruggeman and Westerhoff, 2007). These factors determine the dynamics and structure of the community. It is based upon the identification of processes and patterns engaged in by bacterial species. These microbial interaction patterns are metabolism-driven and responsible for community interaction (Wintermute and Silver, 2010). Social-based microbial interactions (i.e., mutualism, cooperation, and competition, etc.) and the total outcome of these interactions between two microbial populations can be +/+ and −/+ or +/−, respectively (Foster and Bell, 2012). It is said that community structure and function majorly depends on cooperation. The effect of cooperation on community dynamics is determined by engineered cooperation resulting in the synthetic community. Engineered cooperation between two microbial strains (Singh et al., 2016) can be done by manipulation of environmental conditions, i.e., knocking the genes out and in Zuroff et al. (2013). Beyond this, other interaction patterns have been analyzed with engineered microbial species in the synthetic community. Such an application of engineered interaction is highly recognizable in bioremediation strategies (Sharma, 2012). Synthetic biology provides greater potential for the sustainable existence of microorganisms (Dellagnezze et al., 2014) acting together in a large population. Thus, synthetic microbial communities are proved as a key strategy for the bioremediation of contaminants, i.e., pesticides, petroleum (Kachienga et al., 2018), oil spill, acid drainage (Serrano and Leiva, 2017), etc. For building the synthetic microbial communities, the engineered interspecies and intraspecies interactions can make cellular functions robust and enhance the capabilities of microbial consortia in various contaminated scenarios. Quorum sensing is a bacterial signaling mechanism, which is a density-dependent phenomenon via cell-cell communication and population level behavior. The signaling is done by the release and reception of chemical compounds by microbial candidates in a population. This leads to multicellular behavior (Obst, 2007), offering engineerable tasks to design function specific synthetic communities. These synthetic models can also be exploited to obtain a rational design that can lose the function when subjected to competition with other species in the natural environment. With the evolution of genomic constituents and gene transfer, the possibility of the gradual extinction of genetic circuits is present. Thus, strategies are required to maintain the robustness of the synthetic community, achieved via the synthetic models by the development of synergistic and cooperative properties that reduce instability and loss of function (Johns et al., 2016). A recent study by Coyte et al. (2015) suggests that competition among species is significant in determining the stability of communities, acting as a limiting factor in the stability of the synthetic community. Thus, these dynamics must be accelerated in order to design particular function specific synthetic communities for bioremediation purposes (Coyte et al., 2015).
Genetic and Metabolic Engineering
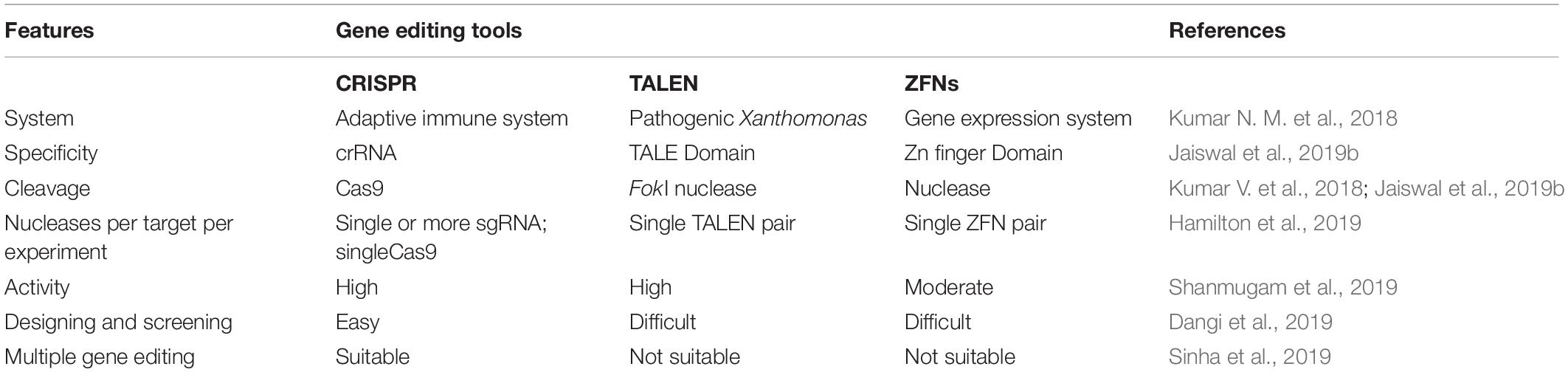
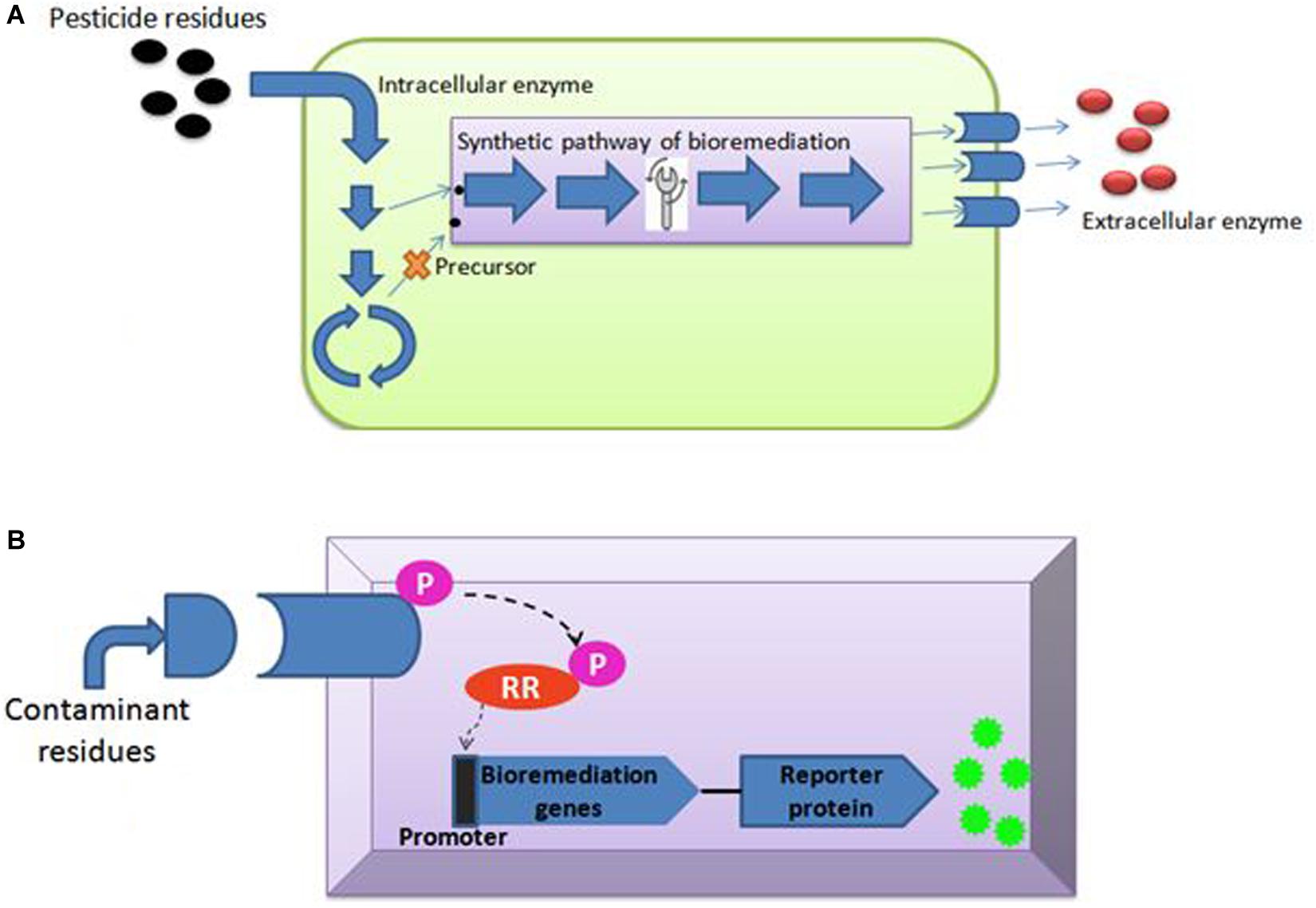
Enríquez (2016) said that genome editing is an umbrella term that refers to “scientific technological advances that enable rational genetic engineering at a local (gene) or global (genome) level to facilitate precise insertion, removal, or substitution of fragments of Deoxyribonucleic acid (“DNA”) molecules, comprising one or more nucleotides into the cell(s) of an organism’s genome.” Transcription-activators like effector nucleases (TALEN), clustered regularly interspaced short palindromic repeats (CRISPR-Cas), and zinc finger nucleases (ZFNs) are major gene editing tools used (Table 2). The most efficient and simple technique of gene editing has been described as CRISPR-Cas (Kanchiswamy et al., 2016). These tools can boost the process of bioremediation. TALEN has a DNA-binding modular which is sequence-specific for the host genome (Utturkar et al., 2013). TALEN binding to DNA creates a double stranded break (DSB) in the target sequence and leaves sticky ends for stability. Similarly, ZFNs is also a DNA-binding domain composed of 30 amino acids. It introduces DSB at the target site of the host genome by the Fok1 cleavage domain. A new sense of using hybrid nucleases containing TALENs and ZFNs nucleases came to act for solving the molecular complications. The CRISPR-Cas system, on the other hand, has unique action properties of high sequence specificity and multiplex gene editing. This unique property is derived from bacteria Streptococcus pyogenes as immunity against the virus. The CRISPR-Cas system consists of crisper derived RNA (crRNA) and trans acting antisense RNA (trcRNA) joined by guide RNA (gRNA). gRNA directs the Cas9 enzyme to introduce DSB in the target DNA sequence by recognizing it. These gene editing tools create the knock-in and knock-out and are under processing for implementation in bioremediation studies (Kumar V. et al., 2018). Recent reports indicate though that the CRISPR-Cas system is mostly adopted and performed by researchers in model organisms i.e., Pseudomonas (Karimi et al., 2015; Nogales et al., 2020) or Escherichia coli (Chen et al., 2018; Marshall et al., 2018; Pontrelli et al., 2018). Nowadays, the new insights toward CRISPR tool kits and designing gRNA for expression of function-specific genes related to remediation in non-model organisms (i.e., Rhodococcus ruber TH, Comamonas testosteroni and Achromobacter sp. HZ01) are also suggested in the field of bioremediation (Mougiakos et al., 2016; Jusiak et al., 2016; Hong et al., 2017; Stein et al., 2018; Tang et al., 2018; Liang et al., 2020). For gene editing and metabolic engineering, the contaminant-inhabited bacteria are the most suitable candidates because they are used to survive and harbor in stress conditions of various toxic, recalcitrant and non-degradable xenobiotics, as discussed above. Moreover, understanding metabolic pathways seems to be important in studying the microbial bioremediation (Plewniak et al., 2018), i.e., bioremediation of toxic pollutants by the haloalkane dehalogenases production pathway and decontamination of pyrethroid from the soil via the biodegradation pathway of fenpropathrin studied in Bacillus sp. DG-02 (Chen et al., 2014). Metabolic engineering leads to modification of the existing pathway for the betterment of the bioremediation process (Michel et al., 2007). This approach majorly covers the study of microbial enzymes, i.e., oxidases, esterases, monooxygenases, oxidoreductases, phenoloxidases involved at various degradation steps (Figure 3A; Mónica and Jaime, 2019; Mujawar et al., 2019). Enzyme-based bioremediation has many advantages because it is an eco-friendly process. The genetic approach increases the perspective of getting recombinant enzymes. There are research reports of extracellular enzymes having a role in enzymatic bioremediation. For instance, arsenic bioremediation (Andres and Bertin, 2016; Choe and Sheppard, 2016; Akhter et al., 2017; Biswas et al., 2019) (bioaccumulation and biotransformation) is achieved via arsenite oxidase coded by aioA gene of Klebsiella pneumonia (Mujawar et al., 2019); enzymes released by white rot fungi degrade PAHs (polycyclic aromatic hydrocarbon) (Zhao and Poh, 2008; Košnár et al., 2019), dyes, TNT (2,4,6- Trinitrotoluene) and PCBs (polychlorinated biphenyls) (Gupte et al., 2016; Kutateladze et al., 2018; Sadańoski et al., 2018). Esterase D enzyme acts on β-endosulfan (organochlorine pesticide), producing simpler compounds (Mehr et al., 2017; Chandra et al., 2019). LiPs encoding hemoproteins from Phanerochaete chrysosporium degrade PAHs. However, incomplete or partial degradation of contaminants lead to simpler non-toxic degradable compounds which can be consumed by microbes (Kumavath and Satyanarayana, 2014) as intermediates in biological pathways or substrate, i.e., LiP (lignin peroxidase) dissimilate benzopyrene to three compounds of quinine, namely 1,6- quinone, 6,12- quinine and 3,6- quinine (Gupta and Pathak, 2020). Furthermore, MnP (Manganese peroxidase) oxidizes organic compounds in the presence of Mn(II) (Xu et al., 2018; Singh et al., 2019). Laccase, MFO (mixed function oxidases), glutathione S transferase, cytochrome P450 also acts in biodegradation of recalcitrant compounds (Singh, 2019; Boudh et al., 2019). Catechol 1,2-dioxygenase (intracellular enzyme) from Pseudomonas NP-6 dissimilate catechol to muconate compounds (Guzik et al., 2011). Also, enzyme immobilization (Cavalca et al., 2013; Sharma B. et al., 2018) increases the half-life, stability, and enzyme activity at a notable level. The enzymatic bioremediation is an elementary, expeditious, and environmental friendly method for microbial removal and degradation of persistent xenobiotics compounds (Sharma B. et al., 2018). Isolation and characterization of microorganisms with enzymatic capabilities have been done with the limitation of less productivity (Rayu et al., 2012). Organophosphates (OP) and organochlorines (OC), major constituents of insecticides, accumulate in the agricultural soil (Panelli et al., 2017) and reach the water bodies via agricultural run-off. Effective bioremediation of γ-hexachlorocyclohexane (OC) and methyl parathion (OP) has been reported by genetically engineered microorganisms (Gong et al., 2016). Moreover, bioremediation of organophosphates and pyrethroids has been experimented with using genetically modified P. putida KT2440 (Zuo et al., 2015). With the advent of metabolic engineering, the catabolism and degradation of various persistent compounds has been reported. The degradation pathways of methyl parathion and γ-hexachlorocyclohexane in Sphingobium japonicum and Pseudomonas sp. WBC-3 witnessed the bioremediation strategy (Liu et al., 2005; Miyazaki et al., 2006). Furthermore, 1, 2, 3-trichloropropane, a persistent constituent of fumigant, is dissimilated into the environment (Techtmann and Hazen, 2016) via heterologous catabolism by the assembly of three enzymes from two different microorganisms in E. coli (Dvorak et al., 2014). A metabolic pathway (Bertin et al., 2011) degrading organophosphorus and paraoxon is engineered by inserting the organophosphorus hydrolase gene (opd) and pnp operon encoding enzymes that convert p-nitrophenol into β-ketoadipate in P. putida (de la Pena Mattozzi et al., 2006). A study showed pobA and chcpca gene clusters of Rhodococcus opacus R7 take part in the bioremediation of naphthenic acid; more specifically, expression aliA1 gene codes for fatty acid CoA ligase for degrading long chains of linear as well as alicyclic naphthenic acid (Zampolli et al., 2020). To minimize the accumulation, the above-mentioned strategy is attained using microbes for partial or complete mineralization of persistent compounds (Miyazaki et al., 2006).
Synthetic Genetic Circuit and Microbial Biosensor
The synthetic genetic circuit requires chassis for implantation. The P. putida is a HVB (Host Vector Biosafety) strain recognized as safe by the Recombinant DNA Advisory Committee. It is also referred to as GRAS (Generally Recognized as Safe) to release in the environment. It is ideal for the next generation of synthetic biology chassis panel because it can withstand high intolerant changing conditions including temperature, pH, solvents, toxins, osmotic, and oxidative stress. Also, P. putida has versatile metabolism and low nutrient requirements (Pabo and Nekludova, 2000). These qualities make this organism the best bacterial model for environmental bioremediation applications (Tanveer et al., 2018). Recently, the P. putida synthetic genetic circuit has been established for the designing of promoter genes and expression of the gene responsible for the degradation of persistent compounds (Adams, 2016). An extension of synthetic biology is the integration of genome with reporter system, and synthetic promoters of P. putida may be developed for synthetic bioremediation pathways. Elmore et al. use serine integrases for synthetic genetic circuit development. Microbial cells have the advantage of a cellular system, which controls cell growth and response to external factors like temperature, light, pH, and oxygen (Tropel and Van Der Meer, 2004). The external environment of microbes inhabiting the contaminated site will respond to concentrations of various persistent compounds present (Ray et al., 2018; Antonacci and Scognamiglio, 2019). Whole cell biosensors for checking the presence, detection and biodegradation potential of xenobiotics compounds (pharmaceutical residues, pesticides, paraffin, PAHs and PCBs, etc.) present (Adhikari, 2019) in environmental samples are attaining attention (Wynn et al., 2017; Heng et al., 2018; Patel et al., 2019). The reporter proteins acting microbe makes a color signal at the detection of particular contaminants via transducer (Zhang and Liu, 2016). A biosensor aiming for detection and bioremediation purposes must have enhanced contact between microbe and contaminant (Dhar et al., 2019). This helps the bacterium to adjust their cellular pathways in response to external environmental conditions and encodes the genes for utilizing the recalcitrant compounds as substrate (Bilal and Iqbal, 2019; Skinder et al., 2020). Synthetic biology strategies are feasible for removing a particular toxic compound because the genetic circuits can be developed against the exogenous environmental toxin (Checa et al., 2012; Tay et al., 2017). The synthetic genetic circuits are assembled via a two-component regulatory system (TCRS) in bacteria (Futagami et al., 2014; Uluşeker et al., 2017). This system acts upon environmental change, and thereby, cells respond to these changes. A prokaryotic TCRS has histidine kinase (HK) and response regulator (RR). The HK is a sensor domain with an extracellular loop present as an integral membrane protein. HK also has a transmitter domain in the last cytoplasmic transmembrane, which is a highly conserved domain. Histidine phosphotransfer (DHp) and catalytic ATP-binding domain (CA) acts for molecular recognition of RR and ATP hydrolysis. The transmitter domain transmits the signal from periplasm to cytoplasm via PAS (Periodic circadian proteins, Aryl hydrocarbon nuclear translocator proteins, and single minded proteins), HAMP (HKs, Adenyltatecyclases, Methyltransferases, and Phosphodiesterases) and GAF (cGMP-specific phosphodiesterases, adenylyl cyclases, and formate hydrogenases) (Casino et al., 2010). Thus, HK senses the external environmental change and adds phosphate to conserved histidine. The HK also regulates RR by phosphorylating the aspartate residues. This promotes the promoter (Figure 3B) binding to activate the gene expression or vice-versa (Ravikumar et al., 2017). Therefore, TCRS-based synthetic biology application for biosensor development for cell-mediated detection and bioremediation can prove to be a new advancement.
Ecological Safety and Risk Assessment
The scientists and researchers are performing the experimental setup to study the bioremediation (Yergeau et al., 2012) potential against various pollutants like oil spill, plastics, synthetic dyes, organic hydrocarbons (Yadav et al., 2015), pesticides (Jaiswal et al., 2019a), heavy metals (Hemmat-Jou et al., 2018; Lebrazi and Fikri-Benbrahim, 2018), and other xenobiotics, etc (Rucká et al., 2017; Paniagua-Michel and Fathepure, 2018; Wu et al., 2020). Considering that bioremediation is performed in an open environment rather than in a closed fermentation tank, the ecological safety of bioremediation performing bacteria must be considered. Economic safety is justified by the metabolic aptness (Gillan et al., 2015) of microorganisms as compared to other traditional physical and chemical bioremediation methods. Besides, regulation for using genetically and metabolically modified bacteria is released to evaluate the possible risks (Khudur et al., 2019). The risk assessment is mainly done by regulatory agencies, i.e., Organization for Economic Cooperation and Development (OECD) at the application level for environmental safety (Russo et al., 2019; Alam and Murad, 2020; Pastor-Jáuregui et al., 2020). The possible risks are gene contamination in the native member of microbial consortium, leading to mislaying of the natural trait (Mills et al., 2019; Pineda et al., 2019; Rycroft et al., 2019). The competitiveness between natural and genetically modified species can give rise to selection pressure on non-target microflora (Kumar N. M. et al., 2018; Mohapatra et al., 2019). Moreover, environment and ecosystem risk assessments infer unpredictable and adverse effects, as discussed above (Cervelli et al., 2016; van Dorst et al., 2020). Particularly, the ecological risk assessment behind addition of GEMs (Genetically Engineered Microorganisms) (Benjamin et al., 2019; Ahankoub et al., 2020) to the native environment rather than a laboratory (Fernandez et al., 2019) is done mainly because of unnecessary delivery of antibiotic resistance marker along with recombinant genome of interest (Davison, 2002; Nora et al., 2019), and unintentional uptake or transfer of induced genes to other microorganisms (French et al., 2019; Janssen and Stucki, 2020). This phenomenon is definitely disturbing the microbial native genome entity (Gangar et al., 2019; Petsas and Vagi, 2019). Another aspect to consider is the change in microbial metabolism (Okino-Delgado et al., 2019), which may release uncertain toxic compounds into the environment, indirectly affecting (negatively) microbial candidates in this context (Myhr and Traavik, 2002). Under the TSCA (Toxic Substances Control Act) (Gardner and Gunsch, 2020), the Office of Pollution Prevention and Toxics (OPPT) programs (Pietro-Souza et al., 2020) of the United States Environmental Protection Agency (Leong and Chang, 2020; Saxena et al., 2020) monitors the environmental and health risks and releases premanufacture legal notice for the accreditation of field research outlines and prototypes (McPartland and McKernan, 2017; Khan et al., 2020). A magnificent example is given by University of Tennessee. In 1995, they gave application and suggested the risk evaluation of microbial bioremediation agents (mainly Pseudomonas fluorescens HK44) on the environment and health (Sayler et al., 1999; Khan et al., 2016; Sharma J. K. et al., 2018; El Zanfaly, 2019). Remarkably, the literature survey points toward a biowar weapon for humanity (Gómez-Tatay and Hernández-Andreu, 2019; Wang and Zhang, 2019), stating that gene editing tools left in bad hands could mislead ethical and moral duties (Khan, 2019; Thakur et al., 2019; Sharma et al., 2020).
Conclusion and Future Perspectives
The microbial bioremediation process for removal and detoxification of contaminants from the environment has now emerged as the best option. Synthetic biology is addressing the decontamination and remediation strategies for xenobiotics and related compounds from the environment. It has been observed that the requisite for understanding existing metabolic pathways is a must for developing synthetic models of bioremediation. Moreover, genomics reconstruction methods (Luo et al., 2014; Marco and Abram, 2019) and technologies made a solid platform for bioremediation studies. Satisfactory progress has been witnessed in the field of bioremediation among various contaminants with the role of specific genes and enzymes applicable via synthetic biology methodologies. Therefore, it is concluded that microbial synthetic biology remediation strategies not only increase the efficiency of microbial bioremediation processes for a particular contaminant, but also provide the best methodologies for researchers.
Author Contributions
SJ contributed to this article under the guidance of PS. SJ acknowledges the support and guidance of PS in pursuing a doctorate under his guidance.
Funding
The author SJ acknowledges Maharshi Dayanand University, Rohtak, India, for providing University Research Scholarship (URS). PS acknowledges the infrastructural support from Department of Science and Technology, New Delhi, Government of India, through FIST grant (Grant No. 1196 SR/FST/LS-I/2017/4).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adams, B. L. (2016). The next generation of synthetic biology chassis: moving synthetic biology from the laboratory to the field. ACS Synth Biol. 5, 1328–1330. doi: 10.1021/acssynbio.6b00256
Adhikari, B. R. (2019). “Advances in the oligonucleotide-based sensor technology for detection of pharmaceutical contaminants in the environment,” in Tools, Techniques and Protocols for Monitoring Environmental Contaminants, eds S. K. Brar, K. Hegde, and V. L. Pachapur (Amsterdam: Elsevier), 125–146.
Agapakis, C. M., Boyle, P. M., and Silver, P. A. (2012). Natural strategies for the spatial optimization of metabolism in synthetic biology. Nat. Chem. Biol. 8, 527–535. doi: 10.1038/nchembio.975
Ahankoub, M., Mardani, G., Ghasemi-Dehkordi, P., Mehri-Ghahfarrokhi, A., Doosti, A., Jami, M. S., et al. (2020). Biodecomposition of phenanthrene and pyrene by a genetically engineered Escherichia coli. Recent Patents Biotechnol. doi: 10.2174/1872208314666200128103513 [Online ahead of print].
Akhter, M., Tasleem, M., Alam, M. M., and Ali, S. (2017). In silico approach for bioremediation of arsenic by structure prediction and docking studies of arsenite oxidase from Pseudomonas stutzeri TS44. International Biodeterioration & Biodegradation 122, 82–91.
Alam, M. M., and Murad, M. W. (2020). The impacts of economic growth, trade openness and technological progress on renewable energy use in organization for economic co-operation and development countries. Renew. Energy 145, 382–390.
Alves, L. D. F., Westmann, C. A., Lovate, G. L., de Siqueira, G. M. V., Borelli, T. C., and Guazzaroni, M. E. (2018). Metagenomic approaches for understanding new concepts in microbial science. Int. J. Genom. 2018:2312987. doi: 10.1155/2018/2312987
Andres, J., and Bertin, P. N. (2016). The microbial genomics of arsenic. FEMS Microbiol. Rev. 40, 299–322. doi: 10.1093/femsre/fuv050
Antonacci, A., and Scognamiglio, V. (2019). Biotechnological advances in the design of algae-based biosensors. Trends Biotechnol. 38, 334–347. doi: 10.1016/j.tibtech.2019.10.005
Arora, P. K., and Bae, H. (2014). Integration of bioinformatics to biodegradation. Biol. Procedures Online 16:8. doi: 10.1186/1480-9222-16-8
Arora, P. K., and Shi, W. (2010). Tools of bioinformatics in biodegradation. Rev. Environ. Sci. Bio/Technol. 9, 211–213.
Atashgahi, S., Hornung, B., Waals, M. J., Rocha, U. N., Hugenholtz, F., Nijsse, B., et al. (2018). A benzene-degrading nitrate-reducing microbial consortium displays aerobic and anaerobic benzene degradation pathways. Sci. Rep. 8:4490. doi: 10.1038/s41598-018-22617-x
Azubuike, C. C., Chikere, C. B., and Okpokwasili, G. C. (2016). Bioremediation techniques–classification based on site of application: principles, advantages, limitations and prospects. World J. Microbiol. Biotechnol. 32:180. doi: 10.1007/s11274-016-2137-x
Banerjee, C., Singh, P. K., and Shukla, P. (2016). Microalgal bioengineering for sustainable energy development: recent transgenesis and metabolic engineering strategies. Biotechnol. J. 11, 303–314. doi: 10.1002/biot.201500284
Bao, Y. J., Xu, Z., Li, Y., Yao, Z., Sun, J., and Song, H. (2017). High-throughput metagenomic analysis of petroleum-contaminated soil microbiome reveals the versatility in xenobiotic aromatics metabolism. J. Environ. Sci. 56, 25–35. doi: 10.1016/j.jes.2016.08.022
Basu, S., Rabara, R. C., Negi, S., and Shukla, P. (2018). Engineering PGPMOs through gene editing and systems biology: a solution for phytoremediation? Trends Biotechnol. 36, 499–510. doi: 10.1016/j.tibtech.2018.01.011
Benjamin, S. R., de Lima, F., and Rathoure, A. K. (2019). “Genetically engineered microorganisms for bioremediation processes: GEMs for bioremediaton,” in Biotechnology: Concepts, Methodologies, Tools, and Applications, Ed. Information Resources Management Association (Pennsylvania: IGI Global), 1607–1634.
Berger, B., Peng, J., and Singh, M. (2013). Computational solutions for omics data. Nat. Rev. Genet. 14:333. doi: 10.1038/nrg3433
Bertin, P. N., Heinrich-Salmeron, A., Pelletier, E., Goulhen-Chollet, F., Arsène-Ploetze, F., Gallien, S., et al. (2011). Metabolic diversity among main microorganisms inside an arsenic-rich ecosystem revealed by meta-and proteo-genomics. ISME J. 5:1735. doi: 10.1038/ismej.2011.51
Bharagava, R. N., Purchase, D., Saxena, G., and Mulla, S. I. (2019). “Applications of metagenomics in microbial bioremediation of pollutants: from genomics to environmental cleanup,” in Microbial Diversity in the Genomic Era, eds S. Das, and H. R. Dash (Cambridge, MA: Academic Press), 459–477.
Biggs, M. B., Medlock, G. L., Kolling, G. L., and Papin, J. A. (2015). Metabolic network modeling of microbial communities. Wiley Interdiscipl. Rev. 7, 317–334. doi: 10.1002/wsbm.1308
Bilal, M., and Iqbal, H. M. (2019). Microbial-derived biosensors for monitoring environmental contaminants: recent advances and future outlook. Process Saf. Environ. Protect. 124, 8–17.
Biswas, R., Vivekanand, V., Saha, A., Ghosh, A., and Sarkar, A. (2019). Arsenite oxidation by a facultative chemolithotrophic Delftia spp. BAs29 for its potential application in groundwater arsenic bioremediation. Int. Biodeteriorat. Biodegrad. 136, 55–62.
Bordbar, A., Monk, J. M., King, Z. A., and Palsson, B. O. (2014). Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 15:107. doi: 10.1038/nrg3643
Bordel, S. (2014). “Genome-scale metabolic models of yeast, methods for their reconstruction, and other applications,” in Yeast Metabolic Engineering, ed. V. Mapelli (New York, NY: Humana Press), 269–279. doi: 10.1007/978-1-4939-0563-8_16
Boudh, S., Singh, J. S., and Chaturvedi, P. (2019). “Microbial resources mediated bioremediation of persistent organic pollutants,” in New and Future Developments in Microbial Biotechnology and Bioengineering, eds J. S. Singh, and D. P. Singh (Amsterdam: Elsevier), 283–294.
Bruggeman, F. J., and Westerhoff, H. V. (2007). The nature of systems biology. TRENDS Microbiol. 15, 45–50.
Bruneel, O., Volant, A., Gallien, S., Chaumande, B., Casiot, C., Carapito, C., et al. (2011). Characterization of the active bacterial community involved in natural attenuation processes in arsenic-rich creek sediments. Microb. Ecol. 61, 793–810. doi: 10.1007/s00248-011-9808-9
Butler, J. E., Young, N. D., and Lovley, D. R. (2010). Evolution of electron transfer out of the cell: comparative genomics of six Geobacter genomes. BMC Genomics 11:40. doi: 10.1186/1471-2164-11-40
Carbajosa, G., and Cases, I. (2010). “Transcriptional networks that regulate hydrocarbon biodegradation,” in Handbook of Hydrocarbon and Lipid Microbiology, eds K. N. Timmis, T. McGenity, J. R. van der Meer, and V. de Lorenzo (Berlin: Springer), 1399–1410.
Casino, P., Rubio, V., and Marina, A. (2010). The mechanism of signal transduction by two-component systems. Curr. Opin. Struct. Biol 20, 763–771. doi: 10.1016/j.sbi.2010.09.010
Cavalca, L., Corsini, A., Zaccheo, P., Andreoni, V., and Muyzer, G. (2013). Microbial transformations of arsenic: perspectives for biological removal of arsenic from water. Future Microbiol. 8, 753–768. doi: 10.2217/fmb.13.38
Cervelli, E., Pindozzi, S., Capolupo, A., Okello, C., Rigillo, M., and Boccia, L. (2016). Ecosystem services and bioremediation of polluted areas. Ecol. Eng. 87, 139–149. doi: 10.1016/j.copbio.2013.10.010
Chakraborty, J., Jana, T., Saha, S., and Dutta, T. K. (2014). R ing-H ydroxylating O xygenase database: a database of bacterial aromatic ring-hydroxylating oxygenases in the management of bioremediation and biocatalysis of aromatic compounds. Environ. Microbiol. Rep. 6, 519–523. doi: 10.1111/1758-2229.12182
Chandra, D., General, T., and Chandra, S. (2019). “Microorganisms: an asset for decontamination of soil,” in Smart Bioremediation Technologies, Ed. P. Bhatt (Cambridge, MA: Academic Press), 319–345.
Checa, S. K., Zurbriggen, M. D., and Soncini, F. C. (2012). Bacterial signaling systems as platforms for rational design of new generations of biosensors. Curr. Opin. Biotechnol 23, 766–772. doi: 10.1016/j.copbio.2012.05.003
Chen, S., Chang, C., Deng, Y., An, S., Dong, Y. H., Zhou, J., et al. (2014). Fenpropathrin biodegradation pathway in Bacillus sp. DG-02 and its potential for bioremediation of pyrethroid-contaminated soils. J. Agricult. Food Chem. 62, 2147–2157. doi: 10.1021/jf404908j
Chen, W., Zhang, Y., Zhang, Y., Pi, Y., Gu, T., Song, L., et al. (2018). CRISPR/Cas9-based genome editing in Pseudomonas aeruginosa and cytidine deaminase-mediated base editing in Pseudomonas species. IScience 6, 222–231. doi: 10.1016/j.isci.2018.07.024
Choe, S. I., and Sheppard, D. C. (2016). “Bioremediation of arsenic using an aspergillus system,” in New and Future Developments in Microbial Biotechnology and Bioengineering, ed. V. K. Gupta (Amsterdam: Elsevier Science), 267–274.
Covert, M. W., Schilling, C. H., Famili, I., Edwards, J. S., Goryanin, I. I., Selkov, E., et al. (2001). Metabolic modeling of microbial strains in silico. Trends Biochem. Sci. 26, 179–186. doi: 10.1016/s0968-0004(00)01754-0
Coyte, K. Z., Schluter, J., and Foster, K. R. (2015). The ecology of the microbiome: networks, competition, and stability. Science 350, 663–666. doi: 10.1126/science.aad2602
Dangi, A. K., Sharma, B., Hill, R. T., and Shukla, P. (2019). Bioremediation through microbes: systems biology and metabolic engineering approach. Crit. Rev. Biotechnol. 39, 79–98. doi: 10.1080/07388551.2018.1500997
Dao, A. T., Vonck, J., Janssens, T. K., Dang, H. T., Brouwer, A., and de Boer, T. E. (2019). Screening white-rot fungi for bioremediation potential of 2, 3, 7, 8-tetrachlorodibenzo-p-dioxin. Indus. Crops Products 128, 153–161.
Das, S., Dash, H. R., and Chakraborty, J. (2016). Genetic basis and importance of metal resistant genes in bacteria for bioremediation of contaminated environments with toxic metal pollutants. Appl. Microbiol. Biotechnol. 100, 2967–2984. doi: 10.1007/s00253-016-7364-4
Davison, J. (2002). Towards safer vectors for the field release of recombinant bacteria. Environ. Biosaf. Res. 1, 9–18. doi: 10.1051/ebr:2002001
De Jong, H. (2002). Modeling and simulation of genetic regulatory systems: a literature review. J. Comput. Biol. 9, 67–103. doi: 10.1089/10665270252833208
de la Pena Mattozzi, M., Tehara, S. K., Hong, T., and Keasling, J. D. (2006). Mineralization of paraoxon and its use as a sole C and P source by a rationally designed catabolic pathway in Pseudomonas putida. Appl. Environ. Microbiol. 72, 6699–6706. doi: 10.1128/AEM.00907-06
Dellagnezze, B. M., de Sousa, G. V., Martins, L. L., Domingos, D. F., Limache, E. E., de Vasconcellos, S. P., et al. (2014). Bioremediation potential of microorganisms derived from petroleum reservoirs. Mar. Pollut. Bull. 89, 191–200. doi: 10.1016/j.marpolbul.2014.10.003
Desai, C., Pathak, H., and Madamwar, D. (2010). Advances in molecular and “-omics” technologies to gauge microbial communities and bioremediation at xenobiotic/anthropogen contaminated sites. Bioresour. Technol. 101, 1558–1569. doi: 10.1016/j.biortech.2009.10.080
Dhar, D., Roy, S., and Nigam, V. K. (2019). “Advances in protein/enzyme-based biosensors for the detection of pharmaceutical contaminants in the environment,” in Tools, Techniques and Protocols for Monitoring Environmental Contaminants, eds S. K. Brar, K. Hegde, and V. L. Pachapur (Amsterdam: Elsevier), 207–229.
Duarte, M., Nielsen, A., Camarinha-Silva, A., Vilchez-Vargas, R., Bruls, T., Wos-Oxley, M. L., et al. (2017). Functional soil metagenomics: elucidation of polycyclic aromatic hydrocarbon degradation potential following 12 years of in situ bioremediation. Environ. Microbiol. 19, 2992–3011. doi: 10.1111/1462-2920.13756
Dvorak, P., Bidmanova, S., Damborsky, J., and Prokop, Z. (2014). Immobilized synthetic pathway for biodegradation of toxic recalcitrant pollutant 1, 2, 3-trichloropropane. Environ. Sci. Technol. 48, 6859–6866. doi: 10.1021/es500396r
Dvořák, P., Nikel, P. I., Damborský, J., and de Lorenzo, V. (2017). Bioremediation 3.0: engineering pollutant-removing bacteria in the times of systemic biology. Biotechnol. Adv. 35, 845–866. doi: 10.1016/j.biotechadv.2017.08.001
El Amrani, A., Dumas, A. S., Wick, L. Y., Yergeau, E., and Berthome, R. (2015). Omics” insights into PAH degradation toward improved green remediation biotechnologies. Environ. Sci. Technol. 49, 11281–11291. doi: 10.1021/acs.est.5b01740
El Zanfaly, H. T. (2019). Biotechnology Contributions in Sustainable Environmental Development. Cairo: National Research Center.
Enríquez, P. (2016). Genome editing and the jurisprudence of scientific empiricism. Vand. J. Ent. & Tech. L. 19:603.
Fajardo, C., Costa, G., Nande, M., Botías, P., García-Cantalejo, J., and Martín, M. (2019). Pb, Cd, and Zn soil contamination: monitoring functional and structural impacts on the microbiome. Appl. Soil Ecol. 135, 56–64.
Fernandez, M., Paisio, C. E., Perotti, R., Pereira, P. P., Agostini, E., and González, P. S. (2019). Laboratory and field microcosms as useful experimental systems to study the bioaugmentation treatment of tannery effluents. J. Environ. Manag. 234, 503–511. doi: 10.1016/j.jenvman.2019.01.019
Foster, K. R., and Bell, T. (2012). Competition, not cooperation, dominates interactions among culturable microbial species. Curr. Biol. 22, 1845–1850. doi: 10.1016/j.cub.2012.08.005
French, K. E., Zhou, Z., and Terry, N. (2019). Horizontal” gene drives” harness indigenous bacteria for bioremediation. bioRxiv [Preprint] doi: 10.1101/735886v2
Futagami, T., Goto, M., and Furukawa, K. (2014). “Genetic system of organohalide-respiring bacteria,” in Biodegradative Bacteria, eds Y. Kamagata, M. Tsuda, and H. Nojiri (Tokyo: Springer), 59–81.
Gangar, T., Bhardwaj, K. K., and Gupta, R. (2019). “Microbes and processes in bioremediation of soil,” in Microbes and Enzymes in Soil Health and Bioremediation, eds A. Kumar, and S. Sharma (Singapore: Springer), 11–37.
Gardner, C. M., and Gunsch, C. K. (2020). “Environmental microbiome analysis and manipulation,” in Women in Water Quality, Ed. D. J. O’Bannon (Cham: Springer), 113–133.
Garg, V., Khan, S., and Dutt, K. (2014). Systematic Analysis of Microbial Degradation Pathway of 1-Naphthyl-N-Methyl carbamate generated by EAWAG Biocatalysis/biodegradation database-pathway prediction System. Int. J. Appl. Sci. Res. Rev. 1, 049–055.
Gillan, D. C., Roosa, S., Kunath, B., Billon, G., and Wattiez, R. (2015). The long-term adaptation of bacterial communities in metal-contaminated sediments: a metaproteogenomic study. Environ. Microbiol. 17, 1991–2005. doi: 10.1111/1462-2920.12627
Gómez-Tatay, L., and Hernández-Andreu, J. M. (2019). Biosafety and biosecurity in synthetic biology: a review. Crit. Rev. Environ. Sci. Technol. 49, 1587–1621.
Gong, J., Liu, X., Cao, X., Diao, Y., Gao, D., Li, H., et al. (2012). PTID: an integrated web resource and computational tool for agrochemical discovery. Bioinformatics 29, 292–294. doi: 10.1093/bioinformatics/bts651
Gong, T., Liu, R., Zuo, Z., Che, Y., Yu, H., Song, C., et al. (2016). Metabolic engineering of Pseudomonas putida KT2440 for complete mineralization of methyl parathion and γ-hexachlorocyclohexane. ACS Synthetic Biol. 5, 434–442. doi: 10.1021/acssynbio.6b00025
Großkopf, T., and Soyer, O. S. (2014). Synthetic microbial communities. Curr. Opin. Microbiol. 18, 72–77. doi: 10.1016/j.mib.2014.02.002
Gupta, S., and Pathak, B. (2020). “Mycoremediation of polycyclic aromatic hydrocarbons,” in Abatement of Environmental Pollutants, eds P. Singh, A. Kumar, and A. Borthakur (Amsterdam: Elsevier), 127–149.
Gupte, A., Tripathi, A., Patel, H., Rudakiya, D., and Gupte, S. (2016). Bioremediation of polycyclic aromatic hydrocarbon (PAHs): a perspective. Open Biotechnol. J. 10, 363–378. doi: 10.1016/j.chemosphere.2013.03.025
Guzik, U., Greń, I., Hupert-Kocurek, K., and Wojcieszyńska, D. (2011). Catechol 1, 2-dioxygenase from the new aromatic compounds–degrading Pseudomonas putida strain N6. Int. Biodeterior. Biodegrad. 65, 504–512.
Hamilton, T. A., Pellegrino, G. M., Therrien, J. A., Ham, D. T., Bartlett, P. C., Karas, B. J., et al. (2019). Efficient inter-species conjugative transfer of a CRISPR nuclease for targeted bacterial killing. Nat. Commun. 10, 1–9. doi: 10.1038/s41467-019-12448-3
Hawley, E. R., Piao, H., Scott, N. M., Malfatti, S., Pagani, I., Huntemann, M., et al. (2014). Metagenomic analysis of microbial consortium from natural crude oil that seeps into the marine ecosystem offshore Southern California. Standards Genom. Sci. 9:1259. doi: 10.4056/sigs.5029016
Hellweger, F. L., Clegg, R. J., Clark, J. R., Plugge, C. M., and Kreft, J. U. (2016). Advancing microbial sciences by individual-based modelling. Nat. Rev. Microbiol. 14:461. doi: 10.1038/nrmicro.2016.62
Hemmat-Jou, M. H., Safari-Sinegani, A. A., Mirzaie-Asl, A., and Tahmourespour, A. (2018). Analysis of microbial communities in heavy metals-contaminated soils using the metagenomic approach. Ecotoxicology 27, 1281–1291. doi: 10.1007/s10646-018-1981-x
Heng, L. Y., Ooi, L., Mori, I. C., and Futra, D. (2018). “Environmental toxicity and evaluation,” in Environmental Risk Analysis for Asian-Oriented, Risk-Based Watershed Management, eds M. Yoneda, and M. Mokhtar (Singapore: Springer), 71–94.
Hong, Y. H., Ye, C. C., Zhou, Q. Z., Wu, X. Y., Yuan, J. P., Peng, J., et al. (2017). Genome sequencing reveals the potential of Achromobacter sp. HZ01 for bioremediation. Front. Microbiol. 8:1507. doi: 10.3389/fmicb.2017.01507
Jacquiod, S., Demanèche, S., Franqueville, L., Ausec, L., Xu, Z., Delmont, T. O., et al. (2014). Characterization of new bacterial catabolic genes and mobile genetic elements by high throughput genetic screening of a soil metagenomic library. J. Biotechnol. 190, 18–29. doi: 10.1016/j.jbiotec.2014.03.036
Jadeja, N. B, More, R. P, Purohit, H. J, and Kapley, A. (2014). Metagenomic analysis of oxygenases from activated sludge. Bioresour. Technol. 165, 250–256. doi: 10.1016/j.biortech.2014.02.045
Jagwani, J., Johnson, J., Datta, M., and Lakshmi, B. J. (2018). Bacterial community dynamics involved in Reactive Orange M2R dye degradation using a real time quantitative PCR and scale up studies using sequence batch reactor. Bioremed. J. 22, 63–71.
Jaiswal, S., Sharma, B., and Shukla, P. (2019a). Integrated approaches in microbial degradation of plastics. Environ. Technol. & Innovat. 17:100567.
Jaiswal, S., Singh, D. K., and Shukla, P. (2019b). Gene editing and systems biology tools for pesticide bioremediation: a review. Front. Microbiol. 10:87. doi: 10.3389/fmicb.2019.00087
Janssen, D. B., and Stucki, G. (2020). Perspectives of genetically engineered microbes for groundwater bioremediation. Environ. Sci. Process. Impacts 22, 487–499. doi: 10.1039/c9em00601j
Johns, N. I., Blazejewski, T., Gomes, A. L., and Wang, H. H. (2016). Principles for designing synthetic microbial communities. Curr. Opin. Microbiol. 31, 146–153. doi: 10.1016/j.mib.2016.03.010
Joshi, G., Schmidt, R., Scow, K. M., Denison, M. S., and Hristova, K. R. (2016). Effect of benzene and ethylbenzene on the transcription of methyl-tert-butyl ether degradation genes of Methylibium petroleiphilum PM1. Microbiology 162, 1563–1571. doi: 10.1099/mic.0.000338
Junghare, M., Spiteller, D., and Schink, B. (2019). Anaerobic degradation of xenobiotic isophthalate by the fermenting bacterium Syntrophorhabdus aromaticivorans. ISME J. 13, 1252–1268. doi: 10.1038/s41396-019-0348-5
Jusiak, B., Cleto, S., Perez-Pińera, P., and Lu, T. K. (2016). Engineering synthetic gene circuits in living cells with CRISPR technology. Trends Biotechnol. 34, 535–547. doi: 10.1016/j.tibtech.2015.12.014
Juwarkar, A. A., Singh, S. K., and Mudhoo, A. (2010). A comprehensive overview of elements in bioremediation. Rev. Environ. Sci. Bio/Technol. 9, 215–288.
Kachienga, L., Jitendra, K., and Momba, M. (2018). Metagenomic profiling for assessing microbial diversity and microbial adaptation to degradation of hydrocarbons in two South African petroleum-contaminated water aquifers. Sci. Rep. 8:7564. doi: 10.1038/s41598-018-25961-0
Kanchiswamy, C. N., Maffei, M., Malnoy, M., Velasco, R., and Kim, J. S. (2016). Fine-tuning next-generation genome editing tools. Trends Biotechnol. 34, 562–574. doi: 10.1016/j.tibtech.2016.03.007
Kapetanovic, I. M. (2008). Computer-aided drug discovery and development (CADDD): in silico-chemico-biological approach. Chem. Biol. Interact 171, 165–176. doi: 10.1016/j.cbi.2006.12.006
Karimi, B., Habibi, M., and Esvand, M. (2015). Biodegradation of naphthalene using Pseudomonas aeruginosa by up flow anoxic–aerobic continuous flow combined bioreactor. J. Environ. Health Sci. Eng. 13, 26. doi: 10.1186/s40201-015-0175-1
Khan, F. I., Husain, T., and Hejazi, R. (2004). An overview and analysis of site remediation technologies. J. Environ. Manag. 71, 95–122. doi: 10.1016/j.jenvman.2004.02.003
Khan, S., Nawab, J., and Waqas, M. (2020). “Constructed wetlands: a clean-green technology for degradation and detoxification of industrial wastewaters,” in Bioremediation of Industrial Waste for Environmental Safety, eds R. N. Bharagava and G. Saxena (Singapore: Springer), 127–163.
Khan, S., Ullah, M. W., Siddique, R., Nabi, G., Manan, S., Yousaf, M., et al. (2016). Role of recombinant DNA technology to improve life. Int. J. Genom. 2016:2405954. doi: 10.1155/2016/2405954
Khan, S. H. (2019). Genome-editing technologies: concept, pros, and cons of various genome-editing techniques and bioethical concerns for clinical application. Mol. Ther. Nucleic Acids 16:326. doi: 10.1016/j.omtn.2019.02.027
Khandelwal, R. A., Olivier, B. G., Röling, W. F., Teusink, B., and Bruggeman, F. J. (2013). Community flux balance analysis for microbial consortia at balanced growth. PLoS One 8:e64567. doi: 10.1371/journal.pone.0064567
Khudur, L. S., Shahsavari, E., Webster, G. T., Nugegoda, D., and Ball, A. S. (2019). The impact of lead co-contamination on ecotoxicity and the bacterial community during the bioremediation of total petroleum hydrocarbon-contaminated soils. Environ. Pollut. 253, 939–948. doi: 10.1016/j.envpol.2019.07.107
Kim, B., Kim, W. J., Kim, D. I., and Lee, S. Y. (2015). Applications of genome-scale metabolic network model in metabolic engineering. J. Indus. Microbiol. Biotechnol. 42, 339–348. doi: 10.1007/s10295-014-1554-9
Kim, S. J., Park, S. J., Cha, I. T., Min, D., Kim, J. S., Chung, W. H., et al. (2014). Metabolic versatility of toluene-degrading, iron-reducing bacteria in tidal flat sediment, characterized by stable isotope probing-based metagenomic analysis. Environ. Microbiol. 16, 189–204. doi: 10.1111/1462-2920.12277
Kondo, R. (2017). “Microbial degradation of endsulfan and endsulfan sulfate,” in Microbe-Induced Degradation of Pesticides, Ed. S. N. Singh (Cham: Springer), 151–166.
Košnár, Z., Èástková, T., Wiesnerová, L., Praus, L., Jablonský, I., Koudela, M., et al. (2019). Comparing the removal of polycyclic aromatic hydrocarbons in soil after different bioremediation approaches in relationto the extracellular enzyme activities. J. Environ. Sci. 76, 249–258. doi: 10.1016/j.jes.2018.05.007
Kües, U. (2015). Fungal enzymes for environmental management. Curr. Opin. Biotechnol 33, 268–278. doi: 10.1016/j.copbio.2015.03.006
Kumar, A., Bisht, B. S., Joshi, V. D., and Dhewa, T. (2011). Review on bioremediation of polluted environment: a management tool. Int. J. Environ. Sci. 1:1079.
Kumar, M., Jaiswal, S., Sodhi, K. K., Shree, P., Singh, D. K., Agrawal, P. K., et al. (2019). Antibiotics bioremediation: perspectives on its ecotoxicity and resistance. Environ. Int. 124, 448–461. doi: 10.1016/j.envint.2018.12.065
Kumar, N. M., Muthukumaran, C., Sharmila, G., and Gurunathan, B. (2018). “Genetically modified organisms and its impact on the enhancement of bioremediation,” in Bioremediation: Applications for Environmental Protection and Management, eds S. J. Varjani, A. K. Agarwal, E. Gnansounou, and B. Gurunathan (Singapore: Springer), 53–76.
Kumar, V., Dangi, A. K., and Shukla, P. (2018). Engineering thermostable microbial xylanases toward its industrial applications. Mol. Biotechnol. 60, 226–235. doi: 10.1007/s12033-018-0059-6
Kumar, P. S. (2019). “Soil bioremediation techniques,” in Advanced Treatment Techniques for Industrial Wastewater, eds H. Athar and A. Sirajuddin (IGI Global), 35–50.
Kumar, S. S., Shantkriti, S., Muruganandham, T., Murugesh, E., Rane, N., and Govindwar, S. P. (2016). Bioinformatics aided microbial approach for bioremediation of wastewater containing textile dyes. Ecol. Inform. 31, 112–121.
Kumavath, R. N., and Satyanarayana, S. V. (2014). Preserving the earth’s microbes for drug discovery in the future. J. Mýcrobýol. Mýcrob. Res. 2, 1–7.
Kutateladze, L., Zakariashvili, N., Khokhashvili, I., Jobava, M., Alexidze, T., Urushadze, T., et al. (2018). Fungal elimination of 2, 4, 6-trinitrotoluene (TNT) from the soils. Eurobiotech J. 2, 39–46.
Lebrazi, S., and Fikri-Benbrahim, K. (2018). “Rhizobium-Legume Symbioses: heavy metal effects and principal approaches for bioremediation of contaminated soil,” in Legumes for Soil Health and Sustainable Management, eds R. S. Meena, A. Das, G. S. Yadav, and R. Lal (Singapore: Springer), 205–233.
Leong, Y. K., and Chang, J. S. (2020). Bioremediation of heavy metals using microalgae: recent advances and mechanisms. Bioresour. Technol. 303:122886. doi: 10.1016/j.biortech.2020.122886
Liang, Y., Jiao, S., Wang, M., Yu, H., and Shen, Z. (2020). A CRISPR/Cas9-based genome editing system for Rhodococcus ruber TH. Metab. Eng. 57, 13–22. doi: 10.1016/j.ymben.2019.10.003
Libis, V., Delepine, B., and Faulon, J. L. (2016a). Expanding biosensing abilities through computer-aided design of metabolic pathways. ACS Synthet. Biol. 5, 1076–1085. doi: 10.1021/acssynbio.5b00225
Libis, V., Delépine, B., and Faulon, J. L. (2016b). Sensing new chemicals with bacterial transcription factors. Curr. Opin. Microbiol. 33, 105–112. doi: 10.1016/j.mib.2016.07.006
Liu, H., Zhang, J. J., Wang, S. J., Zhang, X. E., and Zhou, N. Y. (2005). Plasmid-borne catabolism of methyl parathion and p-nitrophenol in Pseudomonas sp. strain WBC-3. Biochem. Biophys. Res. Commun. 334, 1107–1114. doi: 10.1016/j.bbrc.2005.07.006
Luo, J., Bai, Y., Liang, J., and Qu, J. (2014). Metagenomic approach reveals variation of microbes with arsenic and antimony metabolism genes from highly contaminated soil. PLoS One 9:e108185. doi: 10.1371/journal.pone.0108185
Malla, M. A., Dubey, A., Yadav, S., Kumar, A., Hashem, A., and Abd_Allah, E. F. (2018). Understanding and designing the strategies for the microbe-mediated remediation of environmental contaminants using omics approaches. Front. Microbiol. 9:1132. doi: 10.3389/fmicb.2018.01132
Marco, D. E., and Abram, F. (2019). Using genomics, metagenomics and other” omics” to assess valuable microbial ecosystem services and novel biotechnological applications. Front. Microbiol. 10:151. doi: 10.3389/fmicb.2019.00151
Marshall, R., Maxwell, C. S., Collins, S. P., Jacobsen, T., Luo, M. L., Begemann, M. B., et al. (2018). Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Mol. Cell 69, 146–157. doi: 10.1016/j.molcel.2017.12.007
McPartland, J. M., and McKernan, K. J. (2017). “Contaminants of concern in cannabis: microbes, heavy metals and pesticides,” in Cannabis sativa L.-Botany and Biotechnology, eds S. Chandra, H. Lata, and M. A. ElSohly (Cham: Springer), 457–474.
Mehr, M. A., Farivar, T. N., Najafipour, R., Peymani, A., Alizadeh, S. A., and Johari, P. (2017). Biodegradation of endosulfan as an organochlorine pesticide with Pseudomonas plecoglocissida transfected by LinA gene. Biotechnol. Health Sci. 5:e45306.
Michel, C., Jean, M., Coulon, S., Dictor, M. C., Delorme, F., Morin, D., et al. (2007). Biofilms of As (III)-oxidising bacteria: formation and activity studies for bioremediation process development. Appl. Microbiol. Biotechnol. 77, 457–467. doi: 10.1007/s00253-007-1169-4
Mills, M. G., Ramsden, R., Ma, E. Y., Corrales, J., Kristofco, L. A., Steele, W. B., et al. (2019). CRISPR-generated Nrf2a loss-and gain-of-function mutants facilitate mechanistic analysis of chemical oxidative stress-mediated toxicity in zebrafish. Chem. Res. Toxicol. 33, 426–435. doi: 10.1021/acs.chemrestox.9b00346
Miyazaki, R., Sato, Y., Ito, M., Ohtsubo, Y., Nagata, Y., and Tsuda, M. (2006). Complete nucleotide sequence of an exogenously isolated plasmid, pLB1, involved in γ-hexachlorocyclohexane degradation. Appl. Environ. Microbiol. 72, 6923–6933. doi: 10.1128/AEM.01531-06
Mohanta, T. K., Mohanta, Y. K., and Mohanta, N. (2015). “Role of biotechnology in bioremediation,” in Handbook of Research on Uncovering New Methods for Ecosystem Management Through Bioremediation, eds S. O. Singh and K. Srivastava (Pennsylvania: IGI Global), 399–432.
Mohapatra, B., Kazy, S. K., and Sar, P. (2019). Comparative genome analysis of arsenic reducing, hydrocarbon metabolizing groundwater bacterium Achromobacter sp. KAs 3-5T explains its competitive edge for survival in aquifer environment. Genomics 111, 1604–1619. doi: 10.1016/j.ygeno.2018.11.004
Mónica, M. G. A., and Jaime, M. Q. (2019). “Phenoloxidases of Fungi and Bioremediation,” Fungal Bioremediation: Fundamentals and Applications, eds A. T. Campocosio and H. H. L. Santiesteban (Boca Raton, FL: CRC Press), 62–90.
Mougiakos, I., Bosma, E. F., de Vos, W. M., van Kranenburg, R., and van der Oost, J. (2016). Next generation prokaryotic engineering: the CRISPR-Cas toolkit. Trends Biotechnol. 34, 575–587. doi: 10.1016/j.tibtech.2016.02.004
Mujawar, S. Y., Shamim, K., Vaigankar, D. C., and Dubey, S. K. (2019). Arsenite biotransformation and bioaccumulation by Klebsiella pneumoniae strain SSSW7 possessing arsenite oxidase (aioA) gene. Biometals 32, 65–76. doi: 10.1007/s10534-018-0158-7
Myatt, G. J., Ahlberg, E., Akahori, Y., Allen, D., Amberg, A., Anger, L. T., et al. (2018). In silico toxicology protocols. Regul. Toxicol. Pharmacol. 96, 1–17.
Myhr, A. I., and Traavik, T. (2002). The precautionary principle: Scientific uncertainty and omitted research in the context of GMO use and release. J. Agricult. Environ. Ethics 15, 73–86.
Nogales, J., Mueller, J., Gudmundsson, S., Canalejo, F. J., Duque, E., and Monk, J. (2020). High-quality genome-scale metabolic modelling of Pseudomonas putida highlights its broad metabolic capabilities. Environ. Microbiol. 255–269.
Nora, L. C., Westmann, C. A., Martins-Santana, L., Alves, L. D. F., Monteiro, L. M. O., Guazzaroni, M. E., et al. (2019). The art of vector engineering: towards the construction of next-generation genetic tools. Microb. Biotechnol. 12, 125–147. doi: 10.1111/1751-7915.13318
O’Brien, E. J., Monk, J. M., and Palsson, B. O. (2015). Using genome-scale models to predict biological capabilities. Cell 161, 971–987. doi: 10.1016/j.cell.2015.05.019
Ofaim, S., Zarecki, R., Porob, S., Gat, D., Lahav, T., Xu, X., et al. (2019). Genome-Scale reconstruction of Paenarthrobacter aurescens TC1 metabolic model towards the study of atrazine bioremediation. bioRxiv [Preprint] doi: 10.1101/536011
Okino-Delgado, C. H., Zanutto-Elgui, M. R., do Prado, D. Z., Pereira, M. S., and Fleuri, L. F. (2019). “Enzymatic bioremediation: current status, challenges of obtaining process, and applications,” in Microbial Metabolism of Xenobiotic Compounds, eds P. K. Arora (Singapore: Springer), 79–101.
Orth, J. D., Thiele, I., and Palsson, B. Ø (2010). What is flux balance analysis? Nat. Biotechnol. 28:245. doi: 10.1038/nbt.1614
Pabo, C. O., and Nekludova, L. (2000). Geometric analysis and comparison of protein-DNA interfaces: why is there no simple code for recognition? 1. J. Mol. Biol. 301, 597–624. doi: 10.1006/jmbi.2000.3918
Panelli, S., Capelli, E., Comandatore, F., Landinez-Torres, A., Granata, M. U., Tosi, S., et al. (2017). A metagenomic-based, cross-seasonal picture of fungal consortia associated with Italian soils subjected to different agricultural managements. Fungal Ecol. 30, 1–9.
Paniagua-Michel, J., and Fathepure, B. Z. (2018). “Microbial consortia and biodegradation of petroleum hydrocarbons in marine environments,” in Microbial Action on Hydrocarbons, eds V. Kumar, M. Kumar, and R. Prasad (Singapore: Springer), 1–20.
Panigrahi, S., Velraj, P., and Rao, T. S. (2019). “Functional microbial diversity in contaminated environment and application in bioremediation,” in Microbial Diversity in the Genomic Era, eds S. Das and H. R. Dash (Amsterdam: Academic Press), 359–385.
Pastor-Jáuregui, R., Paniagua-López, M., Martínez-Garzón, J., Martín-Peinado, F., and Sierra-Aragón, M. (2020). Evolution of the residual pollution in soils after bioremediation treatments. Appl. Sci. 10:1006.
Patel, R., Zaveri, P., Mukherjee, A., Agarwal, P. K., More, P., and Munshi, N. S. (2019). Development of fluorescent protein-based biosensing strains: a new tool for the detection of aromatic hydrocarbon pollutants in the environment. Ecotoxicol. Environ. Saf. 182:109450. doi: 10.1016/j.ecoenv.2019.109450
Pavan, M., and Worth, A. P. (2006). Review of QSAR Models for Ready Biodegradation. Brussels: European Commission Directorate General Joint Research Centre.
Peijnenburg, W. J., and Damborský, J. (eds) (2012). Biodegradability Prediction, Vol. 23. Berlin: Springer Science & Business Media.
Petsas, A. S., and Vagi, M. C. (2019). Trends in the bioremediation of pharmaceuticals and other organic contaminants using native or genetically modified microbial strains: a review. Curr. Pharm. Biotechnol 20, 787–824. doi: 10.2174/1389201020666190527113903
Pietro-Souza, W., de Campos Pereira, F., Mello, I. S., Stachack, F. F. F., Terezo, A. J., da Cunha, C. N., et al. (2020). Mercury resistance and bioremediation mediated by endophytic fungi. Chemosphere 240:124874. doi: 10.1016/j.chemosphere.2019.124874
Pineda, M., Lear, A., Collins, J. P., and Kiani, S. (2019). Safe CRISPR: challenges and possible solutions. Trends Biotechnol. 37, 389–401. doi: 10.1016/j.tibtech.2018.09.010
Plewniak, F., Crognale, S., Rossetti, S., and Bertin, P. N. (2018). A genomic outlook on bioremediation: the case of arsenic removal. Front. Microbiol. 9:820. doi: 10.3389/fmicb.2018.00820
Pontrelli, S., Chiu, T. Y., Lan, E. I., Chen, F. Y. H., Chang, P., and Liao, J. C. (2018). Escherichia coli as a host for metabolic engineering. Metab. Eng. 50, 16–46. doi: 10.1016/j.ymben.2018.04.008
Ravikumar, S., Baylon, M. G., Park, S. J., and Choi, J. I. (2017). Engineered microbial biosensors based on bacterial two-component systems as synthetic biotechnology platforms in bioremediation and biorefinery. Microb. Cell Fact. 16:62. doi: 10.1186/s12934-017-0675-z
Rawls, K. D., Dougherty, B. V., Blais, E. M., Stancliffe, E., Kolling, G. L., Vinnakota, K., et al. (2019). A simplified metabolic network reconstruction to promote understanding and development of flux balance analysis tools. Comput. Biol. Med. 105, 64–71. doi: 10.1016/j.compbiomed.2018.12.010
Ray, S., Panjikar, S., and Anand, R. (2018). Design of protein-based biosensors for selective detection of benzene groups of pollutants. ACS Sens. 3, 1632–1638. doi: 10.1021/acssensors.8b00190
Rayu, S., Karpouzas, D. G., and Singh, B. K. (2012). Emerging technologies in bioremediation: constraints and opportunities. Biodegradation 23, 917–926. doi: 10.1007/s10532-012-9576-3
Richelle, A., Gziri, K. M., and Bogaerts, P. (2016). A methodology for building a macroscopic FBA-based dynamical simulator of cell cultures through flux variability analysis. Biochem. Eng. J. 114, 50–64.
Rochfort, S. (2005). Metabolomics reviewed: a new “omics” platform technology for systems biology and implications for natural products research. J. Nat. Prod. 68, 1813–1820. doi: 10.1021/np050255w
Rucká, L., Nešvera, J., and Pátek, M. (2017). Biodegradation of phenol and its derivatives by engineered bacteria: current knowledge and perspectives. World J. Microbiol. Biotechnol. 33:174. doi: 10.1007/s11274-017-2339-x
Russo, F., Ceci, A., Pinzari, F., Siciliano, A., Guida, M., Malusà, E., et al. (2019). Bioremediation of DDT-contaminated agricultural soils: the potential of two autochthonous saprotrophic fungal strains. Appl. Environ. Microbiol. 85, e01720-19. doi: 10.1128/AEM.01720-19
Rycroft, T., Hamilton, K., Haas, C. N., and Linkov, I. (2019). A quantitative risk assessment method for synthetic biology products in the environment. Sci. Total Environ. 696:133940. doi: 10.1016/j.scitotenv.2019.133940
Sadańoski, M. A., Velázquez, J. E., Fonseca, M. I., Zapata, P. D., Levin, L. N., and Villalba, L. L. (2018). Assessing the ability of white-rot fungi to tolerate polychlorinated biphenyls using predictive mycology. Mycology 9, 239–249. doi: 10.1080/21501203.2018.1481152
Salam, L. B., and Ishaq, A. (2019). Biostimulation potentials of corn steep liquor in enhanced hydrocarbon degradation in chronically polluted soil. 3 Biotech 9:46. doi: 10.1007/s13205-019-1580-4
Saxena, G., Purchase, D., and Bharagava, R. N. (2020). “Environmental hazards and toxicity profile of organic and inorganic pollutants of tannery wastewater and bioremediation approaches,” in Bioremediation of Industrial Waste for Environmental Safety, eds R. N. Bharagava and G. Saxena (Singapore: Springer), 381–398. doi: 10.1007/398_2015_5009
Sayler, G. S., Cox, C. D., Burlage, R., Ripp, S., Nivens, D. E., Werner, C., et al. (1999). “Field application of a genetically engineered microorganism for polycyclic aromatic hydrocarbon bioremediation process monitoring and control,” in Novel Approaches for Bioremediation of Organic Pollution, eds S. Reuveny, Y. Flashner, and R. Fass (Boston, MA: Springer), 241–254.
Schloss, P. D., and Handelsman, J. (2008). A statistical toolbox for metagenomics: assessing functional diversity in microbial communities. BMC Bioinformatics 9:34. doi: 10.1186/1471-2105-9-34
Serrano, J., and Leiva, E. (2017). Removal of arsenic using acid/metal-tolerant sulfate reducing bacteria: a new approach for bioremediation of high-arsenic acid mine waters. Water 9:994.
Shah, S. B., Ali, F., Huang, L., Wang, W., Xu, P., and Tang, H. (2018). Complete genome sequence of Bacillus sp. HBCD-sjtu, an efficient HBCD-degrading bacterium. 3 Biotech 8:291. doi: 10.1007/s13205-018-1326-8
Shah, V., Jain, K., Desai, C., and Madamwar, D. (2012). “Molecular analyses of microbial activities involved in bioremediation,” in Microorganisms in Environmental Management, eds A. Prakash and T. Satyanarayana (Dordrecht: Springer), 221–247.
Shanmugam, K., Ramalingam, S., Venkataraman, G., and Hariharan, G. N. (2019). The CRISPR/Cas9 system for targeted genome engineering in free-living fungi: advances and opportunities for lichenized fungi. Front. Microbiol. 10:62. doi: 10.3389/fmicb.2019.00062
Sharma, A., Gupta, G., Ahmad, T., Krishan, K., and Kaur, B. (2020). “Next generation agents (synthetic agents): emerging threats and challenges in detection, protection, and decontamination,” in Handbook on Biological Warfare Preparedness, eds S. J. S. Flora and V. Pachauri (Cambridge, MA: Academic Press), 217–256.
Sharma, B., Dangi, A. K., and Shukla, P. (2018). Contemporary enzyme based technologies for bioremediation: a review. J. Environ. Manag. 210, 10–22. doi: 10.1016/j.jenvman.2017.12.075
Sharma, J. K., Gautam, R. K., Nanekar, S. V., Weber, R., Singh, B. K., Singh, S. K., et al. (2018). Advances and perspective in bioremediation of polychlorinated biphenyl-contaminated soils. Environ. Sci. Pollut. Res. 25, 16355–16375. doi: 10.1007/s11356-017-8995-4
Sharma, S. (2012). Bioremediation: features, strategies and applications. Asian J. Pharm. Life Sci. 2, 202–213.
Shukla, N. (2017). Bioinformatics in environmental bioremediation-a review. Int. J. Sci. Res. Sci. Eng. Technol. 3, 195–205.
Singh, A., and Gothalwal, R. (2018). A reappraisal on biodegradation of fluoride compounds: role of microbes. Water Environ. J. 32, 481–487. doi: 10.1111/wej.12322
Singh, N., Kumar, A., and Sharma, B. (2019). “Role of fungal enzymes for bioremediation of hazardous chemicals,” in Recent Advancement in White Biotechnology Through Fungi, eds A. N. Yadav, S. Mishra, S. Singh, and A. Gupta (Cham: Springer), 237–256.
Singh, N., Srivastava, S., Rathaur, S., and Singh, N. (2016). Assessing the bioremediation potential of arsenic tolerant bacterial strains in rice rhizosphere interface. J. Environ. Sci. 48, 112–119. doi: 10.1016/j.jes.2015.12.034
Singh, S. B. (2018). “Enzyme Catalysis and Its Role in Food Processing Industries,” in Enzymes in Food Technology, Ed. B. A. Law (Singapore: Springer), 143–165.
Singh, V. (2019). “Bioremediation: new prospects for environmental cleaning by enzymes,” in Biotechnology: Concepts, Methodologies, Tools, and Applications, Ed. Information Resources Management Association (Pennsylvania: IGI Global),Google Scholar
Sinha, R., Sharma, B., Dangi, A. K., and Shukla, P. (2019). Recent metabolomics and gene editing approaches for synthesis of microbial secondary metabolites for drug discovery and development. World J. Microbiol. Biotechnol. 35:166. doi: 10.1007/s11274-019-2746-2
Skinder, B. M., Uqab, B., and Ganai, B. A. (2020). “Bioremediation: a sustainable and emerging tool for restoration of polluted aquatic ecosystem,” in Fresh Water Pollution Dynamics and Remediation, eds H. Qadri, R. A. Bhat, M. A. Mehmood, and G. H. Dar (Singapore: Springer), 143–165.
Sridhar, S., and Chandra, J. H. (2014). Involvement of Computational tools towards In Silico remediation-Synthetic textile dyes interacting with Azoreductase. Int. J. Chem. Technol. Res. 6, 4412–4416.
Stein, H. P., Navajas-Pérez, R., and Aranda, E. (2018). “Potential for CRISPR genetic engineering to increase xenobiotic degradation capacities in model fungi,” in Approaches in Bioremediation, eds R. Prasad and E. Aranda (Cham: Springer), 61–78.
Tang, Q., Lu, T., and Liu, S. J. (2018). Developing a synthetic biology toolkit for comamonas testosteroni, an emerging cellular chassis for bioremediation. ACS Synthetic Biol. 7, 1753–1762. doi: 10.1021/acssynbio.7b00430
Tanveer, T., Shaheen, K., Parveen, S., Misbah, Z. T., Babar, M. M., and Gul, A. (2018). “Omics-based bioengineering in environmental biotechnology,” in Omics Technologies and Bio-Engineering, eds D. Barh and V. Azevedo (Cambridge, MA: Academic Press), 353–364.
Tay, P. K. R., Nguyen, P. Q., and Joshi, N. S. (2017). A synthetic circuit for mercury bioremediation using self-assembling functional amyloids. ACS Synthetic Biol. 6, 1841–1850. doi: 10.1021/acssynbio.7b00137
Techtmann, S. M., and Hazen, T. C. (2016). Metagenomic applications in environmental monitoring and bioremediation. J. Indus. Microbiol. Biotechnol. 43, 1345–1354. doi: 10.1007/s10295-016-1809-8
Thakur, M., Medintz, I. L., and Walper, S. A. (2019). Enzymatic bioremediation of organophosphate compounds-progress and remaining challenges. Front. Bioeng. Biotechnol. 7:289. doi: 10.3389/fbioe.2019.00289
Thelusmond, J. R., Strathmann, T. J., and Cupples, A. M. (2019). Carbamazepine, triclocarban and triclosan biodegradation and the phylotypes and functional genes associated with xenobiotic degradation in four agricultural soils. Sci. Total Environ. 657, 1138–1149. doi: 10.1016/j.scitotenv.2018.12.145
Tomei, M. C., and Daugulis, A. J. (2013). Ex situ bioremediation of contaminated soils: an overview of conventional and innovative technologies. Crit. Revi. Environ. Sci. Technol. 43, 2107–2139.
Trigo, A., Valencia, A., and Cases, I. (2008). Systemic approaches to biodegradation. FEMS Microbiol. Rev. 33, 98–108.
Tropel, D., and Van Der Meer, J. R. (2004). Bacterial transcriptional regulators for degradation pathways of aromatic compounds. Microbiol. Mol. Biol. Rev. 68, 474–500.
Uluşeker, C., Torres, J., García, J. L., Hanczyc, M. M., Nogales, J., and Kahramanoğullarý, O. (2017). “September. a dynamic model of the phosphate response system with synthetic promoters in Escherichia coli,” in Proceedings of the Artificial Life Conference, Vol. 14, (Cambridge, MA: MIT Press), 412–419.
Utturkar, S. M., Bollmann, A., Brzoska, R. M., Klingeman, D. M., Epstein, S. E., Palumbo, A. V., et al. (2013). Draft genome sequence for Ralstonia sp. strain OR214, a bacterium with potential for bioremediation. Genome Announc. 1:e00321-13. doi: 10.1128/genomeA.00321-13
van Dorst, J., Wilkins, D., King, C. K., Spedding, T., Hince, G., Zhang, E., et al. (2020). Applying microbial indicators of hydrocarbon toxicity to contaminated sites undergoing bioremediation on subantarctic Macquarie Island. Environ. Pollut. 259:113780. doi: 10.1016/j.envpol.2019.113780
Wang, F., and Zhang, W. (2019). Syntheticbiology: recent progress, biosafety and biosecurity concerns, and possible solutions. J. Biosaf. Biosecur. 1, 22–30.
Wei, K., Yin, H., Peng, H., Lu, G., and Dang, Z. (2019). Bioremediation of triphenyl phosphate in river water microcosms: proteome alteration of Brevibacillus brevis and cytotoxicity assessments. Sci. Total Environ. 649, 563–570. doi: 10.1016/j.scitotenv.2018.08.342
Wintermute, E. H., and Silver, P. A. (2010). Dynamics in the mixed microbial concourse. Genes Dev. 24, 2603–2614. doi: 10.1101/gad.1985210
Wu, Y., Chen, Y., and Wei, N. (2020). Biocatalytic properties of cell surface display laccase for degradation of emerging contaminant acetaminophen in water reclamation. Biotechnol. Bioeng. 117, 342–353. doi: 10.1002/bit.27214
Wynn, D., Deo, S., and Daunert, S. (2017). “Engineering rugged field assays to detect hazardous chemicals using spore-based bacterial biosensors,” in Methods in Enzymology, Vol. 589, eds J. Abelson, M. Simon, G. Verdine, and A. Pyle (Cambridge, MA: Academic Press), 51–85.
Xu, P., Lai, C., Zeng, G., Huang, D., Chen, M., Song, B., et al. (2018). Enhanced bioremediation of 4-nonylphenol and cadmium co-contaminated sediment by composting with Phanerochaete chrysosporium inocula. Bioresour. Technol. 250, 625–634. doi: 10.1016/j.biortech.2017.11.069
Yadav, T. C., Pal, R. R., Shastri, S., Jadeja, N. B., and Kapley, A. (2015). Comparative metagenomics demonstrating different degradative capacity of activated biomass treating hydrocarbon contaminated wastewater. Bioresour. Technol. 188, 24–32. doi: 10.1016/j.biortech.2015.01.141
Yergeau, E., Sanschagrin, S., Beaumier, D., and Greer, C. W. (2012). Metagenomic analysis of the bioremediation of diesel-contaminated Canadian high arctic soils. PLoS One 7:e30058. doi: 10.1371/journal.pone.0030058
Zampolli, J., Di Canito, A., Cappelletti, M., Collina, E., Lasagni, M., and Di Gennaro, P. (2020). Biodegradation of naphthenic acids: identification of Rhodococcus opacus R7 genes as molecular markers for environmental monitoring and their application in slurry microcosms. Appl. Microbiol. Biotechnol. 104, 2675–2689. doi: 10.1007/s00253-020-10378-5
Zhang, D., and Liu, Q. (2016). Biosensors and bioelectronics on smartphone for portable biochemical detection. Biosens. Bioelectron. 75, 273–284. doi: 10.1016/j.bios.2015.08.037
Zhao, B., and Poh, C. L. (2008). Insights into environmental bioremediation by microorganisms through functional genomics and proteomics. Proteomics 8, 874–881. doi: 10.1002/pmic.200701005
Zhu, Y. G., Xue, X. M., Kappler, A., Rosen, B. P., and Meharg, A. A. (2017). Linking genes to microbial biogeochemical cycling: lessons from arsenic. Environ. Sci. Technol. 51, 7326–7339. doi: 10.1021/acs.est.7b00689
Zuo, Z., Gong, T., Che, Y., Liu, R., Xu, P., Jiang, H., et al. (2015). Engineering Pseudomonas putida KT2440 for simultaneous degradation of organophosphates and pyrethroids and its application in bioremediation of soil. Biodegradation 26, 223–233. doi: 10.1007/s10532-015-9729-2
True or False?
Check your Understanding
- Explain bioremediation and the roles that microorganisms can have in this process.
- What is synthetic biology and how can it be applied for bioremediation?
- What are some important considerations and challenges for designing a synthetic microbiome?
- Compare the major techniques for genetic and metabolic engineering.
- How could microbial biosensors contribute to bioremediation efforts?
- What aspects of ecological safety and risk assessment must be addressed for application of synthetic microbiomes for bioremediation in open environments? Why?
Media Attributions
- Video 1 – [microbiome] Microbiome and Planetary Health (6.1) by iMooX at licenced under Creative Commons Attribution-ShareAlike 4.0 International.
- Video 2 – [microbiome] Microbiome and SDGs (6.2) by iMooX at licenced under Creative Commons Attribution-ShareAlike 4.0 International.
References
-
Jaiswal, S., & Shukla, P. (2020). Alternative Strategies for Microbial Remediation of Pollutants via Synthetic Biology. Frontiers in Microbiology, 11. https://www.frontiersin.org/article/10.3389/fmicb.2020.00808

