6 The Oral Microbiome
The Oral Microbiome
The oral microbiome as with other site-specific microbiomes on and inside the human body is very distinct for each individual and its makeup and function is reflective of a variety of factors. Even within the context of the oral cavity, there are several unique niches with their own microbial ecosystem, including saliva, tongue, teeth, gingiva, throat, tonsils, and others. Each of these habitats exhibit diverse and complex interactions between bacteria, archaea, fungi, viruses, and protozoa, where dysfunction can lead to a number diseases, both rare and common (Wade, 2013, Sampaio-Maia et al., 2016).
Factors Affecting Composition
The composition of the oral microbiome and its respective niches is inherently dependent on host genetics, but other factors and habits such diet and smoking have a substantial effect on diversity. Members of the oral cavity demonstrate more heritability than the gut microbiome, and higher abundances of certain heritable organisms, such as Prevotella pallens, are associated with a lack of dental caries, while on the other hand, Streptococcus mutans and Lactobacillus species are linked to cavities (Davenport, 2017, Xiao et al., 2018). Detection of the specific groups of microbes has traditionally been difficult since many are fastidious and cannot be cultivated. So other techniques like metagenomics, metatranscriptomics, and proteomics have been implemented to better characterize the microorganisms present and their respective roles (Parahitiyawa et al., 2010, Grassl et al., 2016). These studies have found that the oral microbiota is quite different even between healthy individuals, and so it may be the microbial functionality that is more important in the progression of various diseases, such as the role of biofilm formation, plaque buildup, and sugar metabolism in the development of dental caries (Segata et al., 2012, Takahashi et al., 2010, Duran-Pinedo and Frias-Lopez, 2015, Sato et al., 2015, Davenport, 2017).
Dysbiosis and Disease
The oral cavity is such a dynamic location that constantly experiences a variety of different foods, drinks, oral hygiene products, and other environmental stimuli with each composed of a multitude of macromolecules, compounds, and potentially other microorganisms, and so it makes sense that its microbiome often fluctuates in diversity (Parahitiyawa et al., 2010). Though there are differences in composition between individuals, specific sites, and over periods of time, species such as Streptococcus mitis and Granulicatella adiacens are conserved and generally present throughout the oral microbiome, while the presence of other specific microbes are associated with a particular disease (Aas et al., 2005). However, diseases like Periodontitis, which is the chronic inflammation of the gums and tooth supporting structures and one of the most common oral diseases, has many organisms associated with the condition, so discerning which ones are primarily responsible is a complex task. There are a variety of viruses that cause oral-related conditions, such as the Human papilloma virus (HPV) which is known for causing lesions and warts in the mouth, as well as head and neck squamous cell carcinoma (Kumaraswamy and Vidhya, 2011). An increase of the protozoa Entamoeba gingivalis and Trichomonas tenax are observed in patients with gingival disease, but are not the causative agents, rather just taking advantage of the increased food sources (bacteria and food debris) from poor hygiene (Wantland et al., 1958). There is also a variety of fungi present in the oral cavity with Candida species being the most common, and many of the members in the oral mycobiome are responsible for chronic diseases, however correlation isn’t exactly clear (Ghannoum et al., 2010). Archaea, primarily methanogens, are also present in the oral microbiome, and while there aren’t technically any known pathogens in this domain, there is an increase in abundance observed in patients with periodontitis (Lepp et al., 2004, Mattarazo et al., 2011, Wade, 2013, Willis and Gabaldon, 2020).
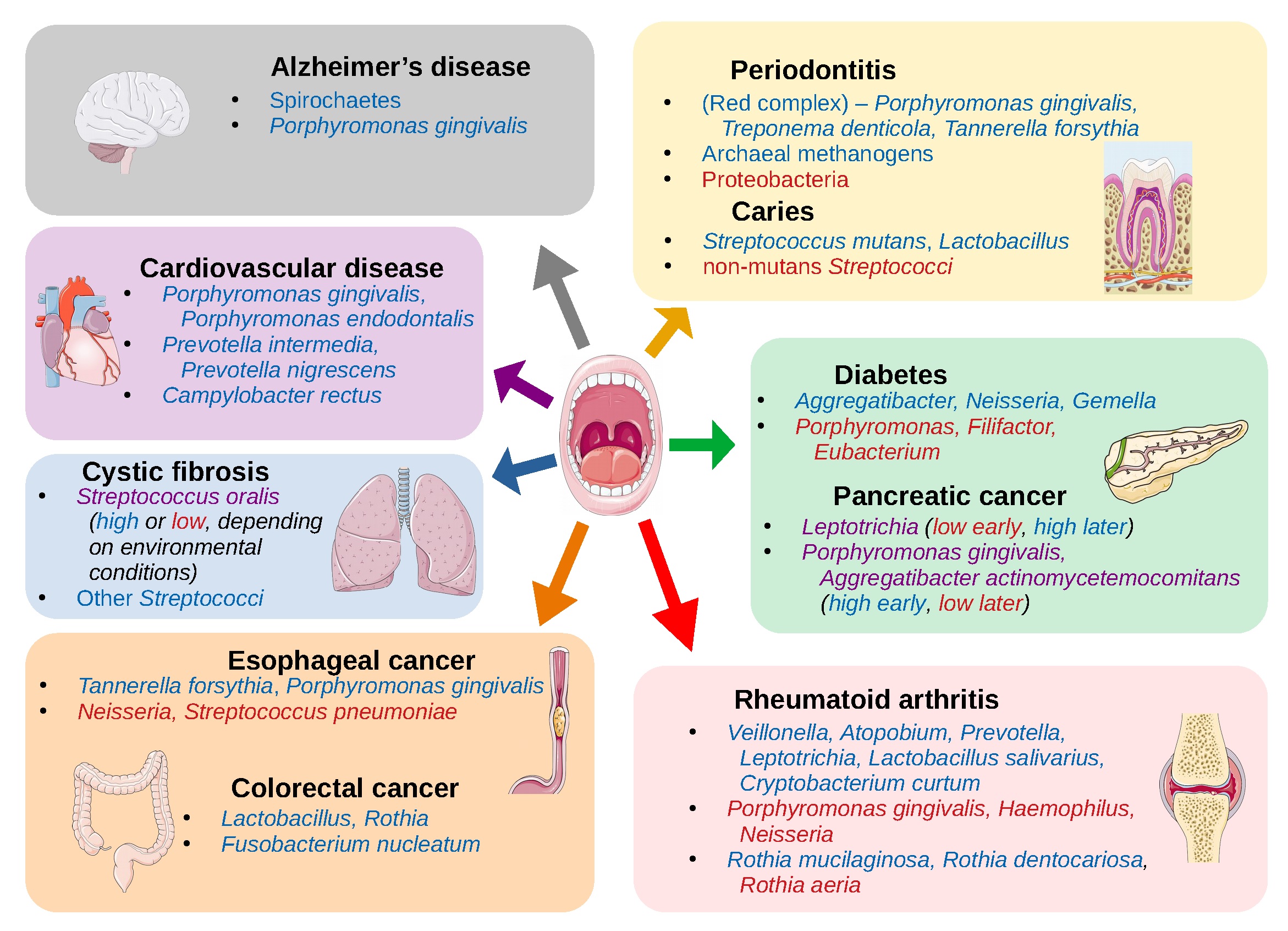
Generally, the oral microbiome is linked with aspects oral health and has primary implications in dental and periodontal diseases, however it can contribute to vitality and diseases in other parts of the body such as cardiovascular disease, stroke, Alzheimer’s disease, cystic fibrosis, rheumatoid arthritis, diabetes, pneumonia, and preterm birth (Seymour et al., 2007, Duran-Pinedo and Frias-Lopez, 2015, Kori et al., 2020, Willis and Gabaldon, 2020). Additionally, the oral microbiome is implicated in various forms of cancer including esophageal, pancreatic, gastric, liver, colorectal, and oral (Willis and Gabaldon, 2020, Bakhti et al., 2021, Mohammed et al., 2021). In many of these types of cancers the Gram-negative anaerobe, Fusobacterium nucleatum, is a primary culprit as it can promote cancer by activation of cell proliferation, promotion of cellular invasion, induction of chronic inflammation and immune evasion (Al-hebshi et al., 2017, McIlvanna et al., 2021). Another contributing factor to cancer development is the use of tobacco products which cause oral microbiome dysbiosis (Al-habshi et al., 2017, Gopinath et al., 2021, Sajid et al., 2021). As with other microbiomes, deviation from the normal composition can result in altered function and progression of disease.
Saliva
Saliva is an important component in maintaining homeostasis in the oral cavity, as it lubricates food, initiates the digestive process, and defends against bacteria. Disruption in secretion can lead to changes in the oral microbiome which promotes progression of oral and other diseases (Grassl et al., 2016). Since saliva contacts virtually all surfaces within the oral cavity it is involved with all other duties of the mouth, and adaptive and innate immune defense mechanisms can be considered the most important in terms of microbiological clinical relevance. The protein and gycoprotein content regulates the oral microbiome by promoting the colonization of commensal microbiota while helping eliminate pathogenic microbes (Cross and Ruhl, 2018). These macromolecules aid in bacterial adhesion and biofilm formation so that they aren’t dislodged by salivary flow and other oral physiological processes (Mandel, 1987). The establishment of beneficial microbes prevents pathogenic bacteria from gaining a foothold, and the agglutinins found in saliva aid in removal through binding and then swallowing (Scannapieco, 1994). The co-evolution of the microbiota and humans has cultivated the development of specific bacterial adhesins for colonization of the preferred microorganisms, thus establishing a mutualism, though pathogenic microbes are quick to adapt to changing binding motifs (Springer and Gagneux, 2013, Cross and Ruhl, 2018).
Saliva can harbor numerous microbes, with one milliliter containing approximately one hundred million microbial cells (Marsh et al., 2015) and over 600 different species (Dewhirst et al., 2020, Willis and Gabaldon, 2020). In one study, the prominent genera found across various types of saliva samples (i.e. spit, drool, and oral rinse) from healthy individuals were Streptococcus (17.5%), Prevotella (15.5%), Veillonella (15.3%), Neisseria (12.7%) and Haemophilus (10%) (Lim et al., 2017). Though it can be difficult to differentiate a core salivary microbiota from other specific oral niches since it coats the oral cavity.
For those suffering from various diseases, the relative abundance of certain microorganisms and general composition of the oral microbiota is altered as compared to healthy controls. For example, patients suffering from chronic obstructive pulmonary disease (COPD) and periodontitis have varying abundances of Veillonella, Rothia, Actinomyces, and Fusobacterium in saliva samples (Lin et al., 2020). Periodontitis and COPD are comorbid diseases that are commonly associated with other conditions like rheumatoid arthritis, diabetes mellitus, and cardiovascular diseases (Scher et al., 2014, Wang et al., 2014, Chrysanthakopoulos and Chrysanthakopoulos, 2014, Lin et al., 2020). Dysfunction of bacterial ecology in saliva is exacerbated by COPD and periodontitis, and so restoration of the salivary microbiota may treat or reduce the severity of these diseases and their comorbidities (Jeffcoat et al., 2014, Zhou et al., 2014, Lin et al., 2020).
Alterations of the salivary microbiome are also associated with certain human viral infections like the herpes virus, influenza, and SARS-CoV (Blostein et al., 2021, Miller et al., 2021). Saliva generally is beneficial to oral health, though changes in it’s makeup could be detrimental and further aggravate disease. For instance, the saliva microbiota and their byproducts could be responsible for increased susceptibility of infection of gum tissues with herpes simplex virus 1, especially in individuals with periodontitis lesions (Zuo et al., 2019). Eukaryotic viruses can also directly interact with oral bacteria and affect disease severity, as is the case with streptococci and influenza which results in an increased viral load (Kamio et al., 2015). Similarly, among patients with COVID-19 there are differences in the salivary bacterial community based on SARS-CoV-2 viral load, though the exact dynamics and repercussions of the interaction isn’t yet well understood. It is possible that COVID-19-induced inflammation could directly impact the oral microbiome and contribute to other diseases connected with dysbiosis (Miller et al., 2021).
Dysbiosis of the fungal component in the salivary microbiome also contributes to overall oral microbial community changes and detrimental effects to the human host. The mycobiome is an important constituent of the oral microbiome though its member’s abundance is much less than that of the bacteriome. There are two main genera in the salivary mycobiome: Candida and Malassezia, where the former is associated with dental plaque bacteria, carbohydrate-rich microbial communities, and acidic pH conditions which contribute to dental caries (Hong et al., 2020). Other interactions between the mycobiome and bacteriome in saliva has been observed in the chronic inflammatory disease oral lichen planus (OLP). OLP causes swelling, discoloration, and open sores of the mucosal membranes in the oral cavity, primarily affecting the buccal region (cheek), but the lips, gingiva, and tongue may also be affected. Similar to plaque buildup and cavity formation, this disease is characterized by an increased abundance of genera Candida, but also Aspergillus, as well as a decrease in biodiversity (Li et al., 2019).
While diversity and abundance of specific groups within the salivary microbiome are associated with various diseases, it is important to consider other factors like diet, lifestyle habits, and genetics that can influence the progression of any particular disease.
Teeth, Plaque, and Cavities
Plaque formation occurs when a multispecies biofilm builds layers on the surface of teeth over time. This structure not only contains a variety of microbes, some of which are pathogenic, but proteins, carbohydrates, minerals, antimicrobial peptides and other compounds that dictate its structure and activity (Amerongen and Veerman, 2002, Flemmig and Beikler, 2011, Zarco et al., 2011). For normal healthy individuals, plaque biofilms are important in maintaining oral homeostasis and good tooth conditions as they can trap pathogens or prevent them from thriving due to competitive inhibition. However, regular detachment of these biofilms through oral hygiene and salivary flow are necessary to prevent pathogen establishment and their escape from immune responses and antimicrobial therapy (Avila et al., 2009, Filoche et al., 2009, Van Essche et al., 2010, Flemmig and Beikler, 2011, Zarco et al., 2011).
The persistence of oral biofilms contributes directly to cavity formation as carbohydrate-fermenting microbes within produce acidic byproducts which lower oral pH and damage tooth enamel (Selwitz et al., 2007, Ling et al., 2010). Cavities form once the surface layers of the tooth wear away and lesions form in the dentin, resulting in oral pain, tooth decay and loss (Selwitz et al., 2007, Zarco et al., 2011). Dental caries are the most prevalent disease for children worldwide, and dental care is the most common unmet need among children in the United States (Loesche and Grenier, 1976, Acs et al., 1999, Low et al., 1999, Peterson et al., 2013).
The fastidious nature of several members of plaque polymicrobial communities have made it traditionally difficult to characterize an exact consortium responsible for dental caries, however, recent studies using NGS technologies have detailed a few signature genera. Streptococcus mutans and Lactobacilli spp. are the primary culprits, but other genera such as Fusobacterium, Bifidobacterium, and Actinomyces are found in high abundance in people with cavities (Munson et al., 2004, Chhour et al., 2005, Corby et al., 2005, Peterson et al., 2013). The fungal yeast, Candida albicans, is also found regularly in children with severe early childhood caries (S-ECC) (Xiao et al., 2018). Interestingly, other members of Streptococcus including S. parasanguinis, S. mitis, S. oralis, and S. sanguinis are associated with individuals exhibiting good dental health (Corby et al., 2005, Peterson et al., 2013). Overall, during the progression of caries there is a reduction in species diversity in these communities (Peterson et al., 2013).
Gingivae and Periodontitis
Aside from dental caries, periodontitis, a.k.a. ‘gum disease’, is the other most common oral disease in humans, and it also results from alterations in oral microbial ecology. This inflammatory condition affects the supporting structures surrounding the teeth where the microbial communities that inhabit the subgingival area serve to trigger its onset (Hajishengallis and Lamont, 2012, Hong et al., 2015). Though progression of the disease comes in episodes, the continual breakdown of periodontium tissue (gingiva, periodontal ligament, cementum, and alveolar bone) leads to alveolar bone breakdown, formation of pocket lesions, and tooth loss (Listgarten, 1986). This condition is very difficult treat as specific antibiotics must be administered, though they are often unsuccessful as pathogens can hide in plaque, develop resistance, and quickly recolonize through reserves in the mucous membranes of the oral cavity. Once pockets form in the periodontium, periodontitis becomes irreversible, as the tissues are unable to reattach to the bone and the pathogens within cannot be completely eradicated or removed (Pihlstrom et al., 2005, Horz and Conrads, 2007, Van Essche et al., 2011, Zarco et al., 2011).
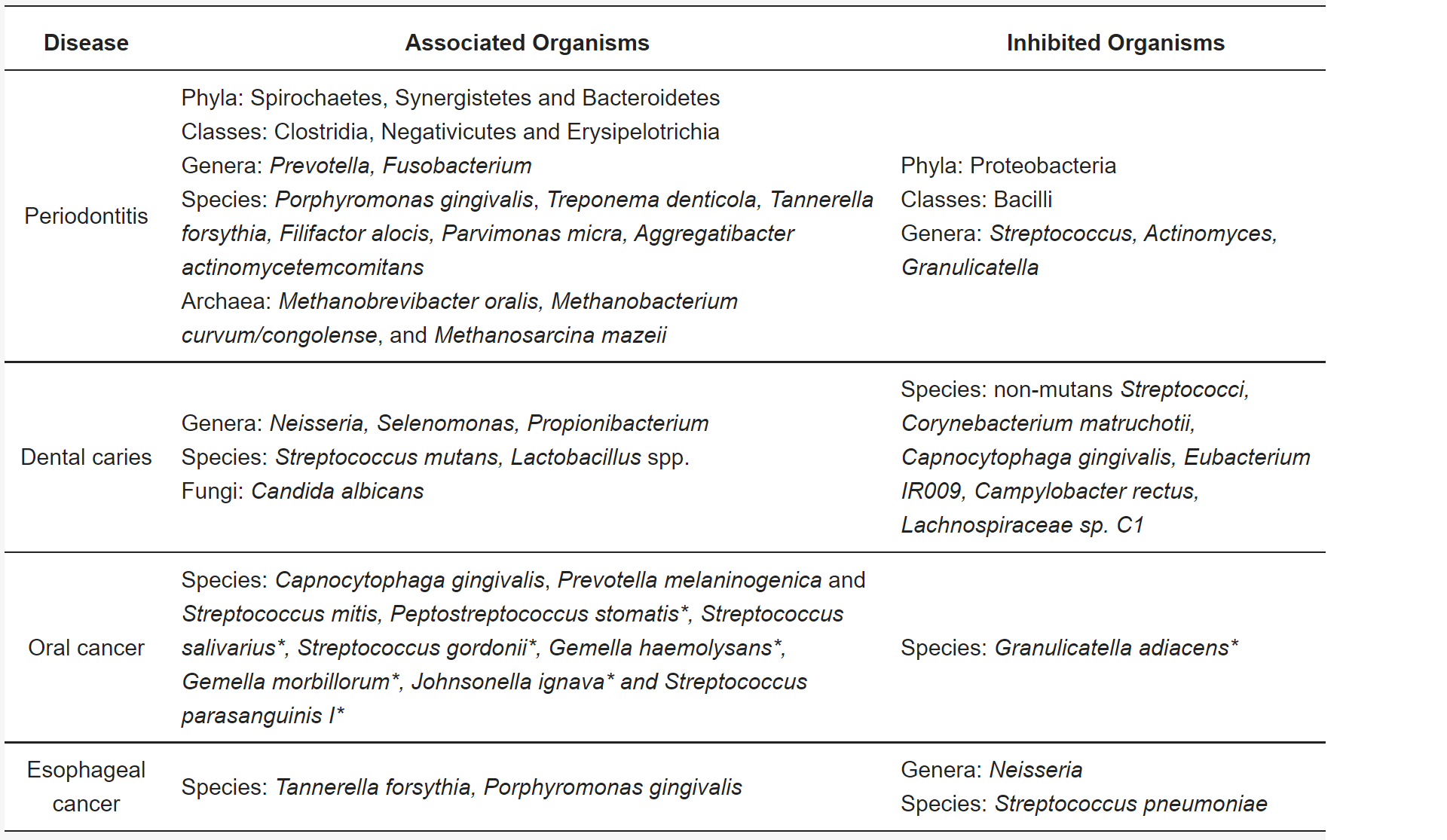
Culture-based studies have shown that periodontitis is associated with varying levels of abundance of three bacterial species: Porphyromonas gingivalis, Tannerella forsythia and Treponema denticola, which are collective referred to as the ‘red complex’ (Socransky et al., 1998, Rocas et al., 2001, Socransky and Hafajee, 2005, Teles et al., 2010). More recent genetic analysis has revealed other bacteria associated with the disease, including those from the classes Clostridia, Negativicutes, and Erysipelotrichia; the genera Synergistes, Prevotella, and Fusobacterium; and the species Filifactor alocis (Vartoukian et al, 2009, Griffen et al., 2012, Costalonga and Herzberg, 2014, Willis and Gabaldon, 2020). Some methanogenic archaeal species have even been implicated in the disease, and perhaps serve a metabolic role as a ‘hydrogen sink’ for secondary fermenters (Lepp et al., 2004, Matarazzo et al., 2011). Viruses, both eukaryotic and bacteriophages, are also thought to play a part in the etiology of periodontitis. While bacteriophages manage biofilm formation through bacterial predation, the role of eukaryotic viruses found in the oral cavity such as those in the Herpesvirus family, is more enigmatic (Martinez et al., 2021). The protist Trichomonas tenax is frequently found in patients with severe periodontitis, though it is not known if its presence is a cause or a result of the disease (Benabdelkader et al., 2019. It is generally thought that this condition arises from events of dysbiosis causing an increase in community diversity and richness which then alters antagonistic/synergistic relationships, metabolic functions, and the oral environment (Kolenbrander et al., 2006, Shi et al., 2015, Ng et al., 2021). Those bacteria that may be absent or inhibited during dysbiosis conditions, and are associated with good periodontal health include those from the phylum Proteobacteria and the Firmicutes, class Bacilli, and the genera Streptococcus, Actinomyces, and Granulicatella (Griffen et al., 2012, Liu et al., 2012, Willis and Gabaldon, 2020).
Periodontitis resulting from oral microbiome dysbiosis has been linked to other diseases like chronic kidney disease, as well as several types of cancers including oral, esophageal, gastric, lung, pancreatic, prostate, hematologic, and breast (Fitzpatrick and Katz, 2010, Ioanndiou and Swede, 2011, Michaud et al., 2017). These associations stemming from the oral cavity could be due to dysbiosis-induced microbiome changes and recruitment of disease causing pathogens, production of harmful microbial-derived byproducts, and/or modulation of the immune system causing an increase in proinflammatory cytokines (Abnet et al., 2005, Kurkivuori et al., 2007, Chalabi et al., 2008, Meurman, 2010, Willis and Gabaldon, 2020). In reality, there are likely a multitude of factors and interactions between microbial communities and the host that lead to the progression of these diseases, however, it is still important to determine certain organismal signatures or functions that could indicate a condition and perhaps aid in diagnosis or treatment.
Tongue
The tongue is another niche for a variety of microbial communities, and though it is a maneuverable oral centerpiece that comes into contact with the rest of the cavity, it has its own unique makeup of microbes. The tongue microbiome serves as an ideal model to analyze changing microbial consortia and understand microbial community dynamics. By employing a novel computational method known as oligotyping, which relies on identifying certain nucleotide content of genetic sequences, a better resolution of microbial taxonomic distribution can be achieved (Eren et al., 2014, Welch et al., 2014, Wilbert et al., 2020). As expected, the tongue experiences large fluctuations in relative abundance across taxa, but this isn’t completely explained by obvious human behavior such as oral hygiene and food/liquid intake. The complex microbial organization that is observed is likely a combination of many factors, like other microbiomes, such as host immunity, physiology influenced by circadian rhythm, epithelial cell renewal, other microbial (e.g. bacteriophage) interactions, as well as certain host behaviors and lifestyles (Welch et al., 2014, Wilbert et al., 2020).
The changing microbiome of the tongue can also be linked to various diseases and conditions in both the oral cavity and throughout the rest of the body. Viewing the tongue for diagnosis isn’t a novel approach either, as traditional Chinese medical practices have viewed tongue phenotypes to discern various illnesses for thousands of years, and currently physical aspects of the tongue are being connected with its microbiome composition to better diagnosis certain diseases like oral, liver, gastric, colorectal, and pancreatic head cancers (Jiang et al., 2012, Han et al., 2014, Mukherjee et al., 2017, Cui et al., 2019 Lu et al., 2019, Mohammed et al., 2021). It is also interesting that members of the tongue microbiome can help regulate blood pressure and cardiovascular health through metabolism of dietary nitrate, and oral hygiene (e.g. tongue cleaning) can help promote the growth of these beneficial organisms (Tribble et al., 2019). While good habits like proper oral hygiene can cultivate a healthy oral microbiome and improve overall health, bad habits like smoking negatively affect microbial communities which contributes to various diseases. For example, metagenomic analysis investigating bacterial species and their gene content showed significant differences of certain species, strains, and metabolic pathways within the tongue microbiome between smokers and never smokers (Sato et al., 2020). This further demonstrates the usefulness of tongue microbiome analysis as a reliable technique to explore and diagnosis certain diseases and conditions.
Conclusion
Though not as well characterized as the gut microbiome, the oral microbiome has a significant impact on human health. Within the oral cavity, specific niche microbiomes of saliva, teeth, gums, and tongue are each compositionally unique and implicated in various conditions and diseases. Moreover still, other oral niches like the buccal mucosa, palate, pharynx, and tonsils not detailed here are distinctive and have certain associated diseases initiated with respective microbiome dysbiosis (Gao et al., 2014, Fukui et al., 2018, van der Meulen et al., 2018). Areas within the oral microbiome may eventually become routine sites for medical observation since samples are easily acquired, especially saliva, and microbial analysis can serve as a fast and reliable option for diagnosis and potential treatment of oral disorders as well as other diseases.
As the oral cavity is the initial point of contact and entrance for foreign microbial loads into the body, oral disease and its associated microbial ecology appear to serve as a conduit for a multitude of other diseases. In a sense the oral microbiome can be considered a precursor to the gut and lung microbiomes, as microbes are undoubtedly mixed with food, drink, and air before being swallowed or inhaled and taken further into the respective system. It is therefore important to understand and better characterize the oral microbiome so that medical diagnosis and intervention can be improved. However, with vast differences in microbial community diversity and composition between humans across the word, this will be quite the challenge to determine normalized healthy consortia respective to people of various geographic regions, ages, lifestlyes, etc.
Hotspot Quiz
Check Your Understanding
- What aspects of the oral microbiome allow for the formation of unique niches?
- How does dysbiosis within the oral microbiome contribute to certain diseases? (What aspects of changes in microbial community diversity and richness could cause oral and other diseases?)
- Why may saliva sampling be considered a viable option for diagnosis of oral and other diseases?
- How do oral biofilms contribute to cavity formation and tooth decay?
- Why are microbial infections associated with periodontitis hard to treat?
- How do certain lifestyles influence various niches of the oral microbiome and lead to disease?
Media Attributions
- Video 1 – GM2: Periodontal Microbiome – Murray Brilliant by National Human Genome Research Institute. Licensed under Creative Commons: By Attribution 3.0 License https://creativecommons.org/licenses/by/3.0/
- Figure 1 – Oral and systemic diseases associated with the oral microbiome by Willis and Gabaldon, 2020, caption adapted by Dylan Parks. Licensed under Creative Commons: By Attribution 4.0 License http://creativecommons.org/licenses/by/4.0/
- Images of body sites and organs in Figure 1 by Servier Medical Art. Licensed under Creative Commons: By Attribution 3.0 License https://creativecommons.org/licenses/by/3.0/
- Figure 2 – Examples of metagenomic studies of associations between the oral microbiome and oral diseases by Willis and Gabaldon, 2020, caption adapted by Dylan Parks. Licensed under Creative Commons: By Attribution 4.0 License http://creativecommons.org/licenses/by/4.0/
References
- Aas, J. A., Paster, B. J., Stokes, L. N., Ingar, O., & Dewhirst, F. E. (2005). Defining the Normal Bacterial Flora of the Oral Cavity. Journal of Clinical Microbiology, 43(11), 5721–5732. https://doi.org/10.1128/JCM.43.11.5721-5732.2005
- Abnet, C. C., Qiao, Y.-L., Dawsey, S. M., Dong, Z.-W., Taylor, P. R., & Mark, S. D. (2005). Tooth loss is associated with increased risk of total death and death from upper gastrointestinal cancer, heart disease, and stroke in a Chinese population-based cohort. International Journal of Epidemiology, 34(2), 467–474. https://doi.org/10.1093/ije/dyh375
- Acs, G., Shulman, R., Ng, M. W., & Chussid, S. (1999). The effect of dental rehabilitation on the body weight of children with early childhood caries. Pediatric dentistry, 21(2), 109–113.
- Al-hebshi, N. N., Alharbi, F. A., Mahri, M., & Chen, T. (2017). Differences in the Bacteriome of Smokeless Tobacco Products with Different Oral Carcinogenicity: Compositional and Predicted Functional Analysis. Genes, 8(4), 106. https://doi.org/10.3390/genes8040106
- Al-hebshi, N. N., Nasher, A. T., Maryoud, M. Y., Homeida, H. E., Chen, T., Idris, A. M., & Johnson, N. W. (2017). Inflammatory bacteriome featuring Fusobacterium nucleatum and Pseudomonas aeruginosa identified in association with oral squamous cell carcinoma. Scientific Reports, 7(1), 1834. https://doi.org/10.1038/s41598-017-02079-3
- Amerongen, A. V. N., & Veerman, E. C. I. (2002). Saliva – the defender of the oral cavity. Oral Diseases, 8(1), 12–22. https://doi.org/10.1034/j.1601-0825.2002.1o816.x
- Avila, M., Ojcius, D. M., & Yilmaz, Ö. (2009). The Oral Microbiota: Living with a Permanent Guest. DNA and Cell Biology, 28(8), 405–411. https://doi.org/10.1089/dna.2009.0874
- Bakhti, S. Z., & Latifi-Navid, S. (2021). Oral microbiota and Helicobacter pylori in gastric carcinogenesis: what do we know and where next? BMC Microbiology, 21(1), 71. https://doi.org/10.1186/s12866-021-02130-4
- Benabdelkader, S., Andreani, J., Gillet, A., Terrer, E., Pignoly, M., Chaudet, H., Aboudharam, G., & la Scola, B. (2019). Specific clones of Trichomonas tenax are associated with periodontitis. PLOS ONE, 14(3), e0213338-. https://doi.org/10.1371/journal.pone.0213338
- Blostein, F., Foote, S., Salzman, E., McNeil, D. W., Marazita, M. L., Martin, E. T., & Foxman, B. (2021). Associations Between Salivary Bacteriome Diversity and Salivary Human Herpesvirus Detection in Early Childhood: A Prospective Cohort Study. Journal of the Pediatric Infectious Diseases Society, 10(8), 856–863. https://doi.org/10.1093/jpids/piab044
- Chalabi, M., Moghim, S., Mogharehabed, A., Najafi, F., & Rezaie, F. (2008). EBV and CMV in chronic periodontitis: a prevalence study. Archives of Virology, 153(10), 1917. https://doi.org/10.1007/s00705-008-0186-7
- Chhour, K. L., Nadkarni, M. A., Byun, R., Martin, F. E., Jacques, N. A., & Hunter, N. (2005). Molecular Analysis of Microbial Diversity in Advanced Caries. Journal of Clinical Microbiology, 43(2), 843–849. https://doi.org/10.1128/JCM.43.2.843-849.2005
- Chrysanthakopoulos, N. A., & Chrysanthakopoulos, P. A. (2016). Association between indices of clinically-defined periodontitis and self-reported history of systemic medical conditions. Journal of Investigative and Clinical Dentistry, 7(1), 27–36. https://doi.org/10.1111/jicd.12119
- Corby, P. M., Lyons-Weiler, J., Bretz, W. A., Hart, T. C., Aas, J. A., Boumenna, T., Goss, J., Corby, A. L., Junior, H. M., Weyant, R. J., & Paster, B. J. (2005). Microbial Risk Indicators of Early Childhood Caries. Journal of Clinical Microbiology, 43(11), 5753–5759. https://doi.org/10.1128/JCM.43.11.5753-5759.2005
- Costalonga, M., & Herzberg, M. C. (2014). The oral microbiome and the immunobiology of periodontal disease and caries. Immunology Letters, 162(2, Part A), 22–38. https://doi.org/10.1016/j.imlet.2014.08.017
- Cross, B. W., & Ruhl, S. (2018). Glycan recognition at the saliva – oral microbiome interface. Cellular Immunology, 333, 19–33. https://doi.org/10.1016/j.cellimm.2018.08.008
- Cui, J., Cui, H., Yang, M., Du, S., Li, J., Li, Y., Liu, L., Zhang, X., & Li, S. (2019). Tongue coating microbiome as a potential biomarker for gastritis including precancerous cascade. Protein & Cell, 10(7), 496–509. https://doi.org/10.1007/s13238-018-0596-6
- Davenport, E. R. (2017). Tooth Be Told, Genetics Influences Oral Microbiome. Cell Host & Microbe, 22(3), 251–253. https://doi.org/https://doi.org/10.1016/j.chom.2017.08.018
- Dewhirst, F. E., Tuste, C., Jacques, Paster, B. J., Tanner, A. C. R., Wen-Han, Y., Abirami, L., & Wade, W. G. (2010). The Human Oral Microbiome. Journal of Bacteriology, 192(19), 5002–5017. https://doi.org/10.1128/JB.00542-10
- Duran-Pinedo, A. E., & Frias-Lopez, J. (2015). Beyond microbial community composition: functional activities of the oral microbiome in health and disease. Microbes and Infection, 17(7), 505–516. https://doi.org/10.1016/j.micinf.2015.03.014
- Eren, A. M., Borisy, G. G., Huse, S. M., & Mark Welch, J. L. (2014). Oligotyping analysis of the human oral microbiome. Proceedings of the National Academy of Sciences, 111(28), E2875. https://doi.org/10.1073/pnas.1409644111
- Filoche, S., Wong, L., & Sissons, C. H. (2009). Oral Biofilms: Emerging Concepts in Microbial Ecology. Journal of Dental Research, 89(1), 8–18. https://doi.org/10.1177/0022034509351812
- Fitzpatrick, S. G., & Katz, J. (2010). The association between periodontal disease and cancer: A review of the literature. Journal of Dentistry, 38(2), 83–95. https://doi.org/10.1016/j.jdent.2009.10.007
- Flemmig, T. F., & Beikler, T. (2011). Control of oral biofilms. Periodontology 2000, 55(1), 9–15. https://doi.org/10.1111/j.1600-0757.2010.00383.x
- Fukui, Y., Aoki, K., Ishii, Y., & Tateda, K. (2018). The palatine tonsil bacteriome, but not the mycobiome, is altered in HIV infection. BMC Microbiology, 18(1), 127. https://doi.org/10.1186/s12866-018-1274-9
- Gao, Z., Kang, Y., Yu, J., & Ren, L. (2014). Human Pharyngeal Microbiome May Play A Protective Role in Respiratory Tract Infections. Genomics, Proteomics & Bioinformatics, 12(3), 144–150. https://doi.org/10.1016/j.gpb.2014.06.001
- Ghannoum, M. A., Jurevic, R. J., Mukherjee, P. K., Cui, F., Sikaroodi, M., Naqvi, A., & Gillevet, P. M. (2010). Characterization of the Oral Fungal Microbiome (Mycobiome) in Healthy Individuals. PLOS Pathogens, 6(1), e1000713-. https://doi.org/10.1371/journal.ppat.1000713
- Gopinath, D., Wie, C. C., Banerjee, M., Thangavelu, L., Kumar R, P., Nallaswamy, D., Botelho, M. G., & Johnson, N. W. (2021). Compositional profile of mucosal bacteriome of smokers and smokeless tobacco users. Clinical Oral Investigations. https://doi.org/10.1007/s00784-021-04137-7
- Grassl, N., Kulak, N. A., Pichler, G., Geyer, P. E., Jung, J., Schubert, S., Sinitcyn, P., Cox, J., & Mann, M. (2016). Ultra-deep and quantitative saliva proteome reveals dynamics of the oral microbiome. Genome Medicine, 8(1), 44. https://doi.org/10.1186/s13073-016-0293-0
- Griffen, A. L., Beall, C. J., Campbell, J. H., Firestone, N. D., Kumar, P. S., Yang, Z. K., Podar, M., & Leys, E. J. (2012). Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. The ISME Journal, 6(6), 1176–1185. https://doi.org/10.1038/ismej.2011.191
- Gross, E. L., Leys, E. J., Gasparovich, E. R., Firestone, N. D., Swartzbaum, J. A., Janies, D. A., Asnani, K., & Griffen, A. L. (2010). Bacterial 16S Sequence Analysis of Severe Caries in Young Permanent Teeth. Journal of Clinical Microbiology, 48(11), 4121–4128. https://doi.org/10.1128/JCM.01232-10
- Hajishengallis, G., & Lamont, R. J. (2012). Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Molecular Oral Microbiology, 27(6), 409–419. https://doi.org/10.1111/j.2041-1014.2012.00663.x
- Han, S., Chen, Y., Hu, J., & Ji, Z. (2014). Tongue images and tongue coating microbiome in patients with colorectal cancer. Microbial Pathogenesis, 77, 1–6. https://doi.org/10.1016/j.micpath.2014.10.003
- Haubek, D. (2010). The highly leukotoxic JP2 clone of Aggregatibacter actinomycetemcomitans: Evolutionary aspects, epidemiology and etiological role in aggressive periodontitis. APMIS, 118.
- Hong, B. Y., Hoare, A., Cardenas, A., Dupuy, A. K., Choquette, L., Salner, A. L., Schauer, P. K., Hegde, U., Peterson, D. E., Dongari-Bagtzoglou, A., Strausbaugh, L. D., & Diaz, P. I. (2020). The Salivary Mycobiome Contains 2 Ecologically Distinct Mycotypes. Journal of Dental Research, 99(6), 730–738. https://doi.org/10.1177/0022034520915879
- Horz, H.-P., & Conrads, G. (2007). Diagnosis and anti-infective therapy of periodontitis. Expert Review of Anti-Infective Therapy, 5(4), 703–715. https://doi.org/10.1586/14787210.5.4.703
- Ioannidou, E., & Swede, H. (2011). Disparities in Periodontitis Prevalence among Chronic Kidney Disease Patients. Journal of Dental Research, 90(6), 730–734. https://doi.org/10.1177/0022034511402209
- Jeffcoat, M. K., Jeffcoat, R. L., Gladowski, P. A., Bramson, J. B., & Blum, J. J. (2014). Impact of Periodontal Therapy on General Health: Evidence from Insurance Data for Five Systemic Conditions. American Journal of Preventive Medicine, 47(2), 166–174. https://doi.org/10.1016/j.amepre.2014.04.001
- Jiang, B., Liang, X., Chen, Y., Ma, T., Liu, L., Li, J., Jiang, R., Chen, T., Zhang, X., & Li, S. (2012). Integrating next-generation sequencing and traditional tongue diagnosis to determine tongue coating microbiome. Scientific Reports, 2(1), 936. https://doi.org/10.1038/srep00936
- Jorth, P., Turner, K. H., Gumus, P., Nizam, N., Buduneli, N., & Whitely, M. (2014). Metatranscriptomics of the Human Oral Microbiome during Health and Disease. MBio, 5(2), e01012-14. https://doi.org/10.1128/mBio.01012-14
- Kamio, N., Imai, K., Shimizu, K., Cueno, M. E., Tamura, M., Saito, Y., & Ochiai, K. (2015). Neuraminidase-producing oral mitis group streptococci potentially contribute to influenza viral infection and reduction in antiviral efficacy of zanamivir. Cellular and Molecular Life Sciences, 72(2), 357–366. https://doi.org/10.1007/s00018-014-1669-1
- Kolenbrander, P. E., Palmer, R. J., Jr, Rickard, A. H., Jakubovics, N. S., Chalmers, N. I., & Diaz, P. I. (2006). Bacterial interactions and successions during plaque development. Periodontology 2000, 42, 47–79. https://doi.org/10.1111/j.1600-0757.2006.00187.x
- Koo, H., & Bowen, W. H. (2014). Candida albicans and Streptococcus mutans: a potential synergistic alliance to cause virulent tooth decay in children. Future Microbiology, 9(12), 1295–1297. https://doi.org/10.2217/fmb.14.92
- Kori, J. A., Saleem, F., Ullah, S., & Azim, M. K. (2020). Characterization of Oral bacteriome dysbiosis in type 2 diabetic patients. MedRxiv, 2020.04.09.20052613. https://doi.org/10.1101/2020.04.09.20052613
- Kumaraswamy, K. L., & Vidya, M. (2011). Human papilloma virus and oral infections: An update. Journal of Cancer Research and Therapeutics, 7(2), 120-127. https://doi.org/10.4103/0973-1482.82915
- Kurkivuori, J., Salaspuro, V., Kaihovaara, P., Kari, K., Rautemaa, R., Grönroos, L., Meurman, J. H., & Salaspuro, M. (2007). Acetaldehyde production from ethanol by oral streptococci. Oral Oncology, 43(2), 181–186. https://doi.org/10.1016/j.oraloncology.2006.02.005
- Lepp, P. W., Brinig, M. M., Ouverney, C. C., Palm, K., Armitage, G. C., & Relman, D. A. (2004). Methanogenic Archaea and human periodontal disease. Proceedings of the National Academy of Sciences of the United States of America, 101(16), 6176. https://doi.org/10.1073/pnas.0308766101
- Li, Y., Wang, K., Zhang, B., Tu, Q., Yao, Y., Cui, B., Ren, B., He, J., Shen, X., van Nostrand, J. D., Zhou, J., Shi, W., Xiao, L., Lu, C., & Zhou, X. (2019). Salivary mycobiome dysbiosis and its potential impact on bacteriome shifts and host immunity in oral lichen planus. International Journal of Oral Science, 11(2), 13. https://doi.org/10.1038/s41368-019-0045-2
- Lim, Y., Totsika, M., Morrison, M., & Punyadeera, C. (2017). The saliva microbiome profiles are minimally affected by collection method or DNA extraction protocols. Scientific Reports, 7(1), 8523. https://doi.org/10.1038/s41598-017-07885-3
- Lin, M., Li, X., Wang, J., Cheng, C., Zhang, T., Han, X., Song, Y., Wang, Z., & Wang, S. (2020). Saliva Microbiome Changes in Patients With Periodontitis With and Without Chronic Obstructive Pulmonary Disease. Frontiers in Cellular and Infection Microbiology, 10, 124. https://www.frontiersin.org/article/10.3389/fcimb.2020.00124
- Ling, Z., Kong, J., Jia, P., Wei, C., Wang, Y., Pan, Z., Huang, W., Li, L., Chen, H., & Xiang, C. (2010). Analysis of Oral Microbiota in Children with Dental Caries by PCR-DGGE and Barcoded Pyrosequencing. Microbial Ecology, 60(3), 677–690. https://doi.org/10.1007/s00248-010-9712-8
- Listgarten, M. A. (1986). Pathogenesis of periodontitis. Journal of Clinical Periodontology, 13(5), 418–425. https://doi.org/10.1111/j.1600-051X.1986.tb01485.x
- Liu, B., Faller, L. L., Klitgord, N., Mazumdar, V., Ghodsi, M., Sommer, D. D., Gibbons, T. R., Treangen, T. J., Chang, Y.-C., Li, S., Stine, O. C., Hasturk, H., Kasif, S., Segrè, D., Pop, M., & Amar, S. (2012). Deep Sequencing of the Oral Microbiome Reveals Signatures of Periodontal Disease. PLOS ONE, 7(6), e37919-. https://doi.org/10.1371/journal.pone.0037919
- Loesche, W. J., & Grenier, E. (1976). Detection of Streptococcus mutans in plaque samples by the direct fluorescent antibody test. Journal of dental research, 55, A87–A93. https://doi.org/10.1177/002203457605500127011
- Low, W., Tan, S., & Schwartz, S. (1999). The effect of severe caries on the quality of life in young children. Pediatric Dentistry, 21(6), 325–326. http://europepmc.org/abstract/MED/10509332
- Lu, H., Ren, Z., Li, A., Li, J., Xu, S., Zhang, H., Jiang, J., Yang, J., Luo, Q., Zhou, K., Zheng, S., & Li, L. (2019). Tongue coating microbiome data distinguish patients with pancreatic head cancer from healthy controls. Journal of Oral Microbiology, 11(1), 1563409. https://doi.org/10.1080/20002297.2018.1563409
- Mager, D. L., Haffajee, A. D., Devlin, P. M., Norris, C. M., Posner, M. R., & Goodson, J. M. (2005). The salivary microbiota as a diagnostic indicator of oral cancer: A descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. Journal of Translational Medicine, 3(1), 27. https://doi.org/10.1186/1479-5876-3-27
- Mandel, I. D. (1987). The Functions of Saliva. Journal of Dental Research, 66(1_suppl), 623–627. https://doi.org/10.1177/00220345870660S103
- Marsh, P. D., Do, T., Beighton, D., & Devine, D. A. (2016). Influence of saliva on the oral microbiota. Periodontology 2000, 70(1), 80–92. https://doi.org/10.1111/prd.12098
- Martínez, A., Kuraji, R., & Kapila, Y. L. (2021). The human oral virome: Shedding light on the dark matter. Periodontology 2000, 87(1), 282–298. https://doi.org/10.1111/prd.12396
- Matarazzo, F., Ribeiro, A. C., Feres, M., Faveri, M., & Mayer, M. P. A. (2011). Diversity and quantitative analysis of Archaea in aggressive periodontitis and periodontally healthy subjects. Journal of Clinical Periodontology, 38(7), 621–627. https://doi.org/10.1111/j.1600-051X.2011.01734.x
- McIlvanna, E., Linden, G. J., Craig, S. G., Lundy, F. T., & James, J. A. (2021). Fusobacterium nucleatum and oral cancer: a critical review. BMC Cancer, 21(1), 1212. https://doi.org/10.1186/s12885-021-08903-4
- Michaud, D. S., Fu, Z., Shi, J., & Chung, M. (2017). Periodontal Disease, Tooth Loss, and Cancer Risk. Epidemiologic Reviews, 39(1), 49–58. https://doi.org/10.1093/epirev/mxx006
- Miller E. H., Annavajhala, M. K., Chong, A. M., Park, H., Nobel, Y. R., Soroush, A., Blackett, J. W., Krigel, A., Phipps, M. M., Freedberg, D. E., Zucker, J., Sano, E. D., Uhlemann, E. C., & Abrams, J. A.(2021). Oral Microbiome Alterations and SARS-CoV-2 Saliva Viral Load in Patients with COVID-19. Microbiology Spectrum, 9(2), e00055-21. https://doi.org/10.1128/Spectrum.00055-21
- Mohammed, M. M. A., al Kawas, S., & Al-Qadhi, G. (2021). Tongue-coating microbiome as a cancer predictor: A scoping review. Archives of Oral Biology, 132, 105271. https://doi.org/10.1016/j.archoralbio.2021.105271
- Mukherjee, P. K., Wang, H., Retuerto, M., Zhang, H., Burkey, B., Ghannoum, M. A., & Eng, C. (2017). Bacteriome and mycobiome associations in oral tongue cancer. Oncotarget, 8(57), 97273–97289. https://doi.org/10.18632/oncotarget.21921
- Munson, M. A., Banerjee, A., Watson, T. F., & Wade, W. G. (2004). Molecular Analysis of the Microflora Associated with Dental Caries. Journal of Clinical Microbiology, 42(7), 3023–3029. https://doi.org/10.1128/JCM.42.7.3023-3029.2004
- Ng, E., Tay, J. R. H., Balan, P., Ong, M. M. A., Bostanci, N., Belibasakis, G. N., & Seneviratne, C. J. (2021). Metagenomic sequencing provides new insights into the subgingival bacteriome and aetiopathology of periodontitis. Journal of Periodontal Research, 56(2), 205–218. https://doi.org/10.1111/jre.12811
- Parahitiyawa, N. B., Scully, C., Leung, W. K., Yam, W. C., Jin, L. J., & Samaranayake, L. P. (2010). Exploring the oral bacterial flora: current status and future directions. Oral Diseases, 16(2), 136–145. https://doi.org/10.1111/j.1601-0825.2009.01607.x
- Peters, B. A., Wu, J., Pei, Z., Yang, L., Purdue, M. P., Freedman, N. D., Jacobs, E. J., Gapstur, S. M., Hayes, R. B., & Ahn, J. (2017). Oral Microbiome Composition Reflects Prospective Risk for Esophageal Cancers. Cancer Research, 77(23), 6777. https://doi.org/10.1158/0008-5472.CAN-17-1296
- Peterson, S. N., Snesrud, E., Liu, J., Ong, A. C., Kilian, M., Schork, N. J., & Bretz, W. (2013). The Dental Plaque Microbiome in Health and Disease. PLOS ONE, 8(3), e58487-. https://doi.org/10.1371/journal.pone.0058487
- Pihlstrom, B. L., Michalowicz, B. S., & Johnson, N. W. (2005). Periodontal diseases. The Lancet, 366(9499), 1809–1820. https://doi.org/10.1016/S0140-6736(05)67728-8
- Pushalkar, S., Ji, X., Li, Y., Estilo, C., Yegnanarayana, R., Singh, B., Li, X., & Saxena, D. (2012). Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiology, 12(1), 144. https://doi.org/10.1186/1471-2180-12-144
- Rôças, I. N., Siqueira, J. F., Jr, Santos, K. R., & Coelho, A. M. (2001). “Red complex” (Bacteroides forsythus, Porphyromonas gingivalis, and Treponema denticola) in endodontic infections: a molecular approach. Oral surgery, oral medicine, oral pathology, oral radiology, and endodontics, 91(4), 468–471. https://doi.org/10.1067/moe.2001.114379
- Sajid, M., Srivastava, S., Joshi, L., & Bharadwaj, M. (2021). Impact of smokeless tobacco-associated bacteriome in oral carcinogenesis. Anaerobe, 70, 102400. https://doi.org/10.1016/j.anaerobe.2021.102400
- Sampaio-Maia, B., Caldas, I. M., Pereira, M. L., Pérez-Mongiovi, D., & Araujo, R. (2016). Chapter Four – The Oral Microbiome in Health and Its Implication in Oral and Systemic Diseases. In S. Sariaslani & G. Michael Gadd (Eds.), Advances in Applied Microbiology (Vol. 97, pp. 171–210). Academic Press. https://doi.org/10.1016/bs.aambs.2016.08.002
- Sato, N., Kakuta, M., Hasegawa, T., Yamaguchi, R., Uchino, E., Kobayashi, W., Sawada, K., Tamura, Y., Tokuda, I., Murashita, K., Nakaji, S., Imoto, S., Yanagita, M., & Okuno, Y. (2020). Metagenomic analysis of bacterial species in tongue microbiome of current and never smokers. Npj Biofilms and Microbiomes, 6(1), 11. https://doi.org/10.1038/s41522-020-0121-6
- Sato, Y., Yamagishi, J., Yamashita, R., Shinozaki, N., Ye, B., Yamada, T., Yamamoto, M., Nagasaki, M., & Tsuboi, A. (2015). Inter-Individual Differences in the Oral Bacteriome Are Greater than Intra-Day Fluctuations in Individuals. PLOS ONE, 10(6), e0131607-. https://doi.org/10.1371/journal.pone.0131607
- Scannapieco, F. A. (1994). Saliva-Bacterium Interactions in Oral Microbial Ecology. Critical Reviews in Oral Biology & Medicine, 5(3), 203–248. https://doi.org/10.1177/10454411940050030201
- Scher, J. U., Bretz, W. A., & Abramson, S. B. (2014). Periodontal disease and subgingival microbiota as contributors for rheumatoid arthritis pathogenesis: modifiable risk factors? Current Opinion in Rheumatology, 26(4), 424–429. https://doi.org/10.1097/BOR.0000000000000076
- Segata, N., Haake, S. K., Mannon, P., Lemon, K. P., Waldron, L., Gevers, D., Huttenhower, C., & Izard, J. (2012). Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biology, 13(6), R42. https://doi.org/10.1186/gb-2012-13-6-r42
- Selwitz, R. H., Ismail, A. I., & Pitts, N. B. (2007). Dental caries. The Lancet, 369(9555), 51–59. https://doi.org/10.1016/S0140-6736(07)60031-2
- Seymour, G. J., Ford, P. J., Cullinan, M. P., Leishman, S., & Yamazaki, K. (2007). Relationship between periodontal infections and systemic disease. Clinical Microbiology and Infection, 13(s4), 3–10. https://doi.org/10.1111/j.1469-0691.2007.01798.x
- Shi, B., Chang, M., Martin, J., Mitreva, M., Lux, R., Klokkevold, P., Sodergren, E., Weinstock, G. M., Haake, S. K., & Li, H. (2021). Dynamic Changes in the Subgingival Microbiome and Their Potential for Diagnosis and Prognosis of Periodontitis. MBio, 6(1), e01926-14. https://doi.org/10.1128/mBio.01926-14
- Socransky, S. S., & Haffajee, A. D. (2005). Periodontal microbial ecology. Periodontology 2000, 38(1), 135–187. https://doi.org/10.1111/j.1600-0757.2005.00107.x
- Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C., & Kent Jr., R. L. (1998). Microbial complexes in subgingival plaque. Journal of Clinical Periodontology, 25(2), 134–144. https://doi.org/10.1111/j.1600-051X.1998.tb02419.x
- Springer, S. A., & Gagneux, P. (2013). Glycan Evolution in Response to Collaboration, Conflict, and Constraint *. Journal of Biological Chemistry, 288(10), 6904–6911. https://doi.org/10.1074/jbc.R112.424523
- Takahashi, N., Washio, J., & Mayanagi, G. (2010). Metabolomics of Supragingival Plaque and Oral Bacteria. Journal of Dental Research, 89(12), 1383–1388. https://doi.org/10.1177/0022034510377792
- Teles, R. P., Gursky, L. C., Faveri, M., Rosa, E. A., Teles, F. R. F., Feres, M., Socransky, S. S., & Haffajee, A. D. (2010). Relationships between subgingival microbiota and GCF biomarkers in generalized aggressive periodontitis. Journal of Clinical Periodontology, 37(4), 313–323. https://doi.org/10.1111/j.1600-051X.2010.01534.x
- Tribble, G. D., Angelov, N., Weltman, R., Wang, B.-Y., Eswaran, S. v, Gay, I. C., Parthasarathy, K., Dao, D.-H. v, Richardson, K. N., Ismail, N. M., Sharina, I. G., Hyde, E. R., Ajami, N. J., Petrosino, J. F., & Bryan, N. S. (2019). Frequency of Tongue Cleaning Impacts the Human Tongue Microbiome Composition and Enterosalivary Circulation of Nitrate. Frontiers in Cellular and Infection Microbiology, 9, 39. https://www.frontiersin.org/article/10.3389/fcimb.2019.00039
- van der Meulen, T. A., Harmsen, H. J. M., Bootsma, H., Liefers, S. C., Vich Vila, A., Zhernakova, A., Fu, J., Wijmenga, C., Spijkervet, F. K. L., Kroese, F. G. M., & Vissink, A. (2018). Dysbiosis of the buccal mucosa microbiome in primary Sjögren’s syndrome patients. Rheumatology, 57(12), 2225–2234. https://doi.org/10.1093/rheumatology/key215
- Van Essche, M., Quirynen, M., Sliepen, I., Loozen, G., Boon, N., van Eldere, J., & Teughels, W. (2011). Killing of anaerobic pathogens by predatory bacteria. Molecular Oral Microbiology, 26(1), 52–61. https://doi.org/10.1111/j.2041-1014.2010.00595.x
- Vartoukian, S. R., Palmer, R. M., & Wade, W. G. (2009). Diversity and Morphology of Members of the Phylum “Synergistetes” in Periodontal Health and Disease. Applied and Environmental Microbiology, 75(11), 3777–3786. https://doi.org/10.1128/AEM.02763-08
- Wade, W. G. (2013). The oral microbiome in health and disease. Pharmacological Research, 69(1), 137–143. https://doi.org/10.1016/j.phrs.2012.11.006
- Wang, L., Ganly, I. (2014). The oral microbiome and oral cancer. Clinics in Laboratory Medicine, 34, 711–719. https://doi.org/10.1016/j.cll.2014.08.004
- Wang, T.-F., Jen, I.-A., Chou, C., & Lei, Y.-P. (2014). Effects of periodontal therapy on metabolic control in patients with type 2 diabetes mellitus and periodontal disease: a meta-analysis. Medicine, 93(28), e292–e292. https://doi.org/10.1097/MD.0000000000000292
- Wantland, W. W., Wantland, E. M., Remo, J. W., & Winquist, D. L. (1958). Studies on Human Mouth Protozoa. Journal of Dental Research, 37(5), 949–950. https://doi.org/10.1177/00220345580370052601
- Welch, J. L., Utter, D. R., Rossetti, B. J., Mark Welch, D. B., Eren, A. M., & Borisy, G. G. (2014). Dynamics of tongue microbial communities with single-nucleotide resolution using oligotyping. Frontiers in Microbiology, 5, 568. https://www.frontiersin.org/article/10.3389/fmicb.2014.00568
- Wilbert, S. A., Mark Welch, J. L., & Borisy, G. G. (2020). Spatial Ecology of the Human Tongue Dorsum Microbiome. Cell Reports, 30(12), 4003-4015.e3. https://doi.org/10.1016/j.celrep.2020.02.097
- Willis, J. R., & Gabaldón, T. (2020). The Human Oral Microbiome in Health and Disease: From Sequences to Ecosystems. Microorganisms, 8(2). https://doi.org/10.3390/microorganisms8020308
- Xiao, J., Grier, A., Faustoferri, R. C., Alzoubi, S., Gill, A. L., Feng, C., Liu, Y., Quivey, R. G., Kopycka-Kedzierawski, D. T., Koo, H., & Gill, S. R. (2018). Association between Oral Candida and Bacteriome in Children with Severe ECC. Journal of Dental Research, 97(13), 1468–1476. https://doi.org/10.1177/0022034518790941
- Zarco, M. F., Vess, T. J., & Ginsburg, G. S. (2012). The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Diseases, 18(2), 109–120. https://doi.org/10.1111/j.1601-0825.2011.01851.x
- Zhou, X., Han, J., Liu, Z., Song, Y., Wang, Z., & Sun, Z. (2014). Effects of periodontal treatment on lung function and exacerbation frequency in patients with chronic obstructive pulmonary disease and chronic periodontitis: A 2-year pilot randomized controlled trial. Journal of Clinical Periodontology, 41(6), 564–572. https://doi.org/https://doi.org/10.1111/jcpe.12247
- Zuo, Y., Whitbeck, J. C., Haila, G. J., Hakim, A. A., Rothlauf, P. W., Eisenberg, R. J., Cohen, G. H., & Krummenacher, C. (2019). Saliva enhances infection of gingival fibroblasts by herpes simplex virus 1. PLOS ONE, 14(10), e0223299-. https://doi.org/10.1371/journal.pone.0223299

