8 The Respiratory Microbiome
The Respiratory Microbiome
The respiratory microbiome (specifically the lower portions) was once thought to be nonexistent, as many considered healthy lungs to be a sterile environment. Like other microbiomes though, the upper and lower respiratory system are environments rich with bacteria, fungi, archaea, and viruses, and because of the novelty of study many of the members in these groups have not been identified. However, it is known that many of such organisms are not only responsible for or exacerbate a number of pulmonary diseases, but that the respiratory microbiome can help to mitigate infection and influence treatment of certain illnesses (Unger and Bogaert, 2017, Watson et al., 2019).
The human respiratory system begins with the upper respiratory system; starting with the nose and nostrils, then proceeds to the nasal cavity, pharynx, and larynx. The lower respiratory system then starts with the trachea, moves to the bronchi and bronchioles, and ends with the lungs. Each of these structures and areas have their own microbial niche which serve various roles in maintaining health, but also have the potential to be compromised. The respiratory system is similar to the oral and digestive system, in that they interact intimately with external stimuli and bring in foreign matter and microbes during regular biological processes, like inhalation. These systems also have overlapping parts (e.g. oral cavity and pharynx), where their respective microbiomes serve similar roles such as resistance to disruptive environmental factors and host defense (Dickson et al., 2014, Zaura et al., 2014, Hakansson et al., 2018). The mucosal surfaces of the respiratory tract also have a natural healthy biofilm where the resident microbiota reside to maintain homeostatic functions, however, as an extension of dysbiosis, these biofilms can become altered and obtain pathogenic microbes which contribute to respiratory diseases (Hamilos, 2019).
Composition
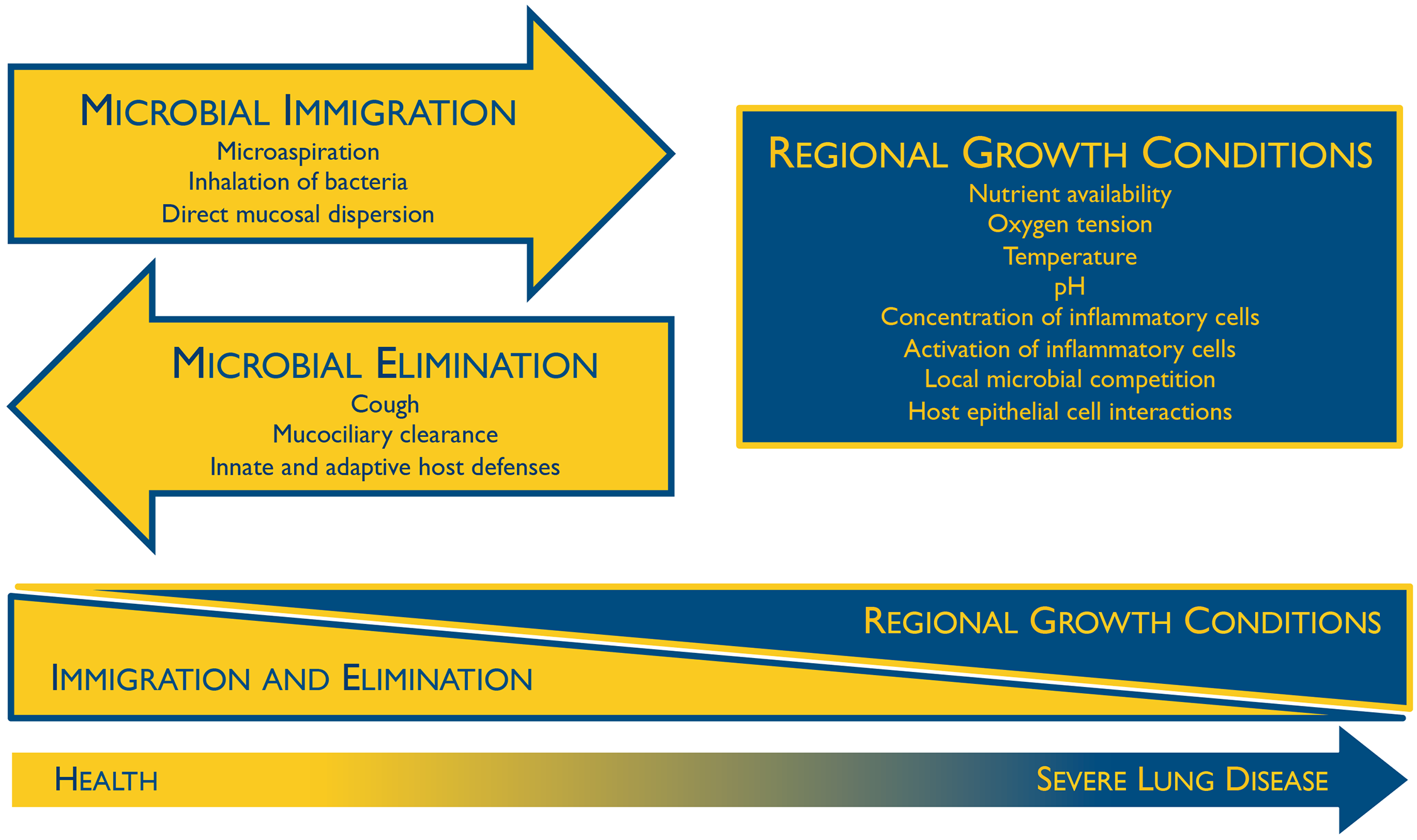
Immediately after birth the respiratory microbiome becomes colonized, though the collection of microbes found here during infancy is not much different than other locations in the body, and generally reflect the mode of delivery (Dominguez-Bello et al., 2010). The respiratory microbiome, as well as others, become differentiated over time when the different species of microbes adapt and outcompete each other based on their niche, host genetics, and environmental factors (Bosch et al., 2016). Colonization additionally depends on microbial immigration, microbial elimination, and relative reproduction rates of its members (Figure 1) (Dickson and Huffnagle, 2015). With age, the respiratory microbiome of each particular individual reflects the aforementioned factors as well as other lifestyle habits, and so they can be vastly different between people. Although these respiratory microbiomes are unique, a core set of microbes can be defined, with most of them being aerobic or facultatively anaerobic (Stearns et al., 2015, Hakansson et al., 2018).
Microbial composition also depends on the specific structure or niche within the respiratory system. The nasal microbiome largely consists of Staphylococci, Corynebacteria, and Streptococci, which is most likely due to its similarity and proximity to the skin (Mika et al., 2015, Shilts et al., 2016). Communities within the paranasal sinuses are highly diverse and include lactic-acid producing bacteria such as Lactobacilli, Enterococcus, and Pediococcus, and once in the nasopharynx the collection of microbes becomes more complex and favors oxygen-utilizing groups (Abreu et al., 2012, Biesbroek et. al, 2014, Teo et al., 2015). In the lower respiratory system, the microbiome resembles a bit of a mix from both the nasal and oral cavities, with a majority of bacteria from the genera Streptococcus, Fusobacteria, Pseudomonas, Veillonella, and Prevotella (Cui et al., 2014, Beck et al., 2015, Dickson and Huffnagle, 2015, Hakansson et al., 2018).
While much focus of the respiratory microbiome has been on bacteria (like many other microbiomes), study of the mycobiome and virome components have revealed their importance in pulmonary health and disease. Fungal spores are commonly inhaled, especially in high numbers during peak seasons, and depending on the particular organism and host immune health, they can cause infection and respiratory complications (Pashley et al., 2012, Denning et al., 2014, Nguyen et al., 2015). For healthy people, the respiratory mycobiome is predominated by environmental fungi including Aspergillus, Cladosporium, and Penicillium species (Charlson et al., 2012, van Woerden et al., 2012, Underhill and Iliev, 2014). However, members from these genera and other more well-known fungal pathogens, like Candida albicans, can be implicated in respiratory illnesses due to microbial community changes (Nguyen et al., 2015). The respiratory virome can also modulate various pulmonary diseases through respective bacterial interactions and host immune response. For bacteriophage composition, there are observed differences between healthy individuals and patients suffering from diseases associated with respective bacterial infections (Wilner et al., 2009). There is a diverse range of eukaryotic viruses that make up the normal human and respiratory virome, with the majority consisting of adenoviruses, herpesviruses and human papillomaviruses (HPVs), though these may only be transient and quickly cleared by the immune system or via the mucociliary escalator (Wilner et al., 2009, Popgeorgiev et al., 2013). More research needs to be conducted over the normal respiratory virome to determine its functionality and importance, though usually more focus is catered to those viruses directly involved in pathogenesis and dysbiosis.
Dysbiosis and Disease
The normal matrix of organisms in the respiratory system maintain homeostasis through their complex interactive network with each other and their host. Pathogens like Streptococcus pneumoniae and Haemophilus influenzae are actually part of the resident respiratory microbiome and interact with commensal microbes, however, they can incite infection when they are not kept in check due to community perturbation (de Steenhuijsen et al., 2015). During events of dysbiosis, pathogens and opportunistic pathogens can contribute to and exacerbate several respiratory diseases including asthma, chronic obstructive pulmonary disease (COPD), cystic fibrosis, pneumonia, otitis media, and other acute infections like influenza.
Asthma is a chronic lung disease that usually develops at an early age, and its complex etiology makes it difficult to treat. People with asthma experience inflammation and tightening of their airways and a production of extra mucus impairs breathing, talking, and being active. The prevalence of asthma has increased in recent decades, and a decrease in microbe exposure during youth could be partially responsible (Ober and Yao, 2011, Prescott, 2013). Proper development of the immune system is reliant on contact with microbes, and those individuals who are regularly subjected to a diverse variety of microorganisms at an early age have a reduced risk of developing asthma (Ownby et al., 2002, Fujimura et al., 2010, Fall et al., 2015). Aside from grooming the immune system, the intake of microbes influences the composition of the mucosal microbiota that lines the air passageways, which could directly affect asthma development (Durack et al., 2016). Indeed epidemiologic studies have shown that the disruption of the microbiome, such as in the case of antibiotic use during childhood, create a predisposition for this allergic disease (Khalkhali et al., 2014). Specifically, the increased presence of genus Haemophilus and other Proteobacteria, as well as the fungus Aspergillus fumigatus in the respiratory system are signature of asthma patients (Hilty et al., 2010, Teo et al., 2015, Urb et al., 2015). These pathogens would normally be cleared by the immune system or stifled by the resident microbiota, however, early childhood dysbiosis resulting in hampered immune functions and/or a discontent microbial consortium in the respiratory system contribute to the development of this condition.
Chronic obstructive pulmonary disease (COPD) is another chronic inflammatory lung disease that causes obstructed airflow to the lungs and is exacerbated by microbial dysbiosis. Emphysema, a condition where the alveoli are damaged due to cigarette smoke and other irritants, and chronic bronchitis, a condition characterized by inflammation of the lining of the airways, are the most common contributors to COPD. An alteration of the lower respiratory microbiome causing reduced diversity or the presence of specific organisms can further aggravate COPD causing a worsening of symptoms (Huang et al., 2014). Viral infections, like those caused by the rhinovirus, are commonly detected (in about half the patients) during COPD exacerbations (Seemungal et al., 2001, Rohde et al., 2003, Mallia et al., 2011). Also during these bouts, there is an increase in pathogens from the bacterial genera Haemophilus, Pseudomonas, and Moraxella and a general shift towards the phylum Proteobacteria in the respiratory microbiome (Huang et al., 2014, Millares et al., 2014). Additionally, patients with COPD exhibit a decline or absence of certain normal lung organisms that could contribute to mucosal homeostasis and prevention of pathogen overgrowth, such as Firmicutes (Streptococcus spp.), as well as Bacteroidetes (Prevotella spp.) (Fukata and Arditi, 2014, Sze et al., 2015, Hakansson et al., 2018). These compositional changes are at least partially brought about by current COPD treatments like steroid and antibiotic usage, and so future therapy may need to take the respiratory microbiome into further consideration for various approaches (Wang et al., 2016).
Cystic fibrosis (CF) is a genetic disease that causes normal cellular secretion of mucus, sweat, and digestive juices to become thicker and viscous. In the respiratory system, the buildup of sticky mucus can clog airways, impair lung function, and make breathing difficult. Bacterial pathogens, like Staphylococcus aureus, Pseudomonas aeruginosa, and Haemophilus parainfluenzae, tend to grow in the sputum and can cause infections which worsening CF conditions (Keogh and Stanojevic, 2018). Their biofilm-forming capabilities also contribute to the excess mucus production, further causing complications as infectious agents which can be resilient and persistent (Høiby et al, 2010, Orazi and O’Toole, 2017). While changes in the microbiome are not typically responsible for exacerbations of CF, antibiotic administration is the primary treatment option to eliminate these pathogens residing in the mucus, however, prolonged use can result in decreased microbiome diversity and dysbiosis in the respiratory system as well as other areas (Zhao et al., 2012, Carmody et al., 2013 Price et al., 2013, Dickson et al., 2014).
Pneumonia is a microbial infection of the lungs that can cause the alveoli to fill with pus or other liquids, and severe cases can be life-threatening, especially to children, the elderly, and those who are immunocompromised (Wardlaw et al., 2006, Chalmers et al., 2011, Valley et al., 2015). The intensity of the infection is directly correlated with the microbial load and diversity, so etiologic determination is important for proper treatment, as the onset of pneumonia can have bacterial, fungal, or viral origins (Dickson et al., 2014, Iwai et al., 2014). Viral pneumonia is commonly caused by influenza or rhinovirus, however, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has now also become a major cause post 2019 (Jain et al., 2015, Zhou et al., 2020, Pettigrew et al., 2021). Respiratory viral infections are the most common cause acute respiratory illnesses, and often times, these viral infections then lead to secondary bacterial infections, like pneumonia, due to host immune reactions and induced dysbiosis in the respiratory tract and gastrointestinal system (Hanada et al., 2018). Streptococcus pneumoniae is the most common culprit for bacterial pneumonia, and while it regularly resides in the upper respiratory tract, under dysbiotic conditions this organism can proliferate and spread to the lower respiratory tract and cause illness (File, 2003, de Steenhuijsen Piters et al., 2014). In patients with pneumonia, there is a decrease in many Gram-negative anaerobic bacteria that are apart of the resident microbiota, some of which are associated with a reduced risk of hospital-acquired pneumonia and clearance of S. pneumoniae (Bousbia et al., 2012, de Steenhuijsen Piters et al., 2014, Krone et al., 2014). Although, changes in the respiratory microbiome are more apparent in bacterial pneumonia rather than viral, it is not known whether this alteration in microbial consortia is a cause or effect of the disease, though it is likely conditional of both circumstances (Ramos-Sevillano et al., 2019, Pettigrew et al., 2021).
Conclusion
The respiratory microbiome is an up-and-coming avenue for research, diagnosis, and treatment of several pulmonary illnesses. While much of its characterization is still in it’s infancy, proper identification of microorganisms is important for management of respiratory diseases (Nguyen et al., 2016). As antibiotic resistance continues to surge, novel approaches are necessary to mitigate not only respiratory illnesses, but to maintain general health. And similarly to other microbiome-related treatments, like fecal microbiota transplant for the gut, eventually oral or aerosol administered microbiota may be implemented to treat pulmonary conditions (Huang and Boushey, 2015). Continual research detailing both core and individual respiratory microbiomes will push the advancement of these ‘futuristic’ medical motions.
Drag and Drop Quiz
Drag each contributing factor of the respiratory microbiome composition into the appropriate category.
Check your Understanding
- How is the colonization of the respiratory microbiome influenced, and what factors affect its composition over time?
- What factors of the respiratory microbiome affect the development of asthma?
- Which microorganisms are implicated in COPD and periods of exacerbation? How does dysbiosis contribute to this chronic condition?
- Why do you think a greater change is observed in the respiratory microbiota in patients with bacterial pneumonia versus viral?
Media Attributions
- Figure 1 – Ecological Determinants of the Respiratory Microbiome by Dickson and Huffnagle, 2015. Licensed under Creative Commons: Attribution 4.0 International (CC BY 4.0) License https://creativecommons.org/licenses/by/4.0/
- Video 1 – The Lung Microbiome: Challenging Old Paradigms about Microbes by National Human Genome Research Institute. Licensed under Creative Commons: By Attribution 3.0 License https://creativecommons.org/licenses/by/3.0/
References
- Abreu, N. A., Nagalingam, N. A., Song, Y., Roediger, F. C., Pletcher, S. D., Goldberg, A. N., & Lynch, S. V. (2012). Sinus Microbiome Diversity Depletion and Corynebacterium tuberculostearicum Enrichment Mediates Rhinosinusitis. Science Translational Medicine, 4(151), 151ra124-151ra124. https://doi.org/10.1126/scitranslmed.3003783
- Beck, J. M., Schloss, P. D., Venkataraman, A., Twigg, H., Jablonski, K. A., Bushman, F. D., Campbell, T. B., Charlson, E. S., Collman, R. G., Crothers, K., Curtis, J. L., Drews, K. L., Flores, S. C., Fontenot, A. P., Foulkes, M. A., Frank, I., Ghedin, E., Huang, L., Lynch, S. v, … Young, V. B. (2015). Multicenter Comparison of Lung and Oral Microbiomes of HIV-infected and HIV-uninfected Individuals. American Journal of Respiratory and Critical Care Medicine, 192(11), 1335–1344. https://doi.org/10.1164/rccm.201501-0128OC
- Biesbroek, G., Tsivtsivadze, E., Sanders, E. A. M., Montijn, R., Veenhoven, R. H., Keijser, B. J. F., & Bogaert, D. (2014). Early Respiratory Microbiota Composition Determines Bacterial Succession Patterns and Respiratory Health in Children. American Journal of Respiratory and Critical Care Medicine, 190(11), 1283–1292. https://doi.org/10.1164/rccm.201407-1240OC
- Bosch, A. A. T. M., Levin, E., van Houten, M. A., Hasrat, R., Kalkman, G., Biesbroek, G., de Steenhuijsen Piters, W. A. A., de Groot, P.-K. C. M., Pernet, P., Keijser, B. J. F., Sanders, E. A. M., & Bogaert, D. (2016). Development of Upper Respiratory Tract Microbiota in Infancy is Affected by Mode of Delivery. EBioMedicine, 9, 336–345. https://doi.org/10.1016/j.ebiom.2016.05.031
- Bousbia, S., Papazian, L., Saux, P., Forel, J. M., Auffray, J.-P., Martin, C., Raoult, D., & la Scola, B. (2012). Repertoire of Intensive Care Unit Pneumonia Microbiota. PLOS ONE, 7(2), e32486-. https://doi.org/10.1371/journal.pone.0032486
- Carmody, L. A., Zhao, J., Schloss, P. D., Petrosino, J. F., Murray, S., Young, V. B., Li, J. Z., & LiPuma, J. J. (2013). Changes in Cystic Fibrosis Airway Microbiota at Pulmonary Exacerbation. Annals of the American Thoracic Society, 10(3), 179–187. https://doi.org/10.1513/AnnalsATS.201211-107OC
- Chalmers, J. D., Taylor, J. K., Mandal, P., Choudhury, G., Singanayagam, A., Akram, A. R., & Hill, A. T. (2011). Validation of the Infectious Diseases Society of America/American Thoratic Society Minor Criteria for Intensive Care Unit Admission in Community-Acquired Pneumonia Patients Without Major Criteria or Contraindications to Intensive Care Unit Care. Clinical Infectious Diseases, 53(6), 503–511. https://doi.org/10.1093/cid/cir463
- Charlson, E. S., Diamond, J. M., Bittinger, K., Fitzgerald, A. S., Yadav, A., Haas, A. R., Bushman, F. D., & Collman, R. G. (2012). Lung-enriched Organisms and Aberrant Bacterial and Fungal Respiratory Microbiota after Lung Transplant. American Journal of Respiratory and Critical Care Medicine, 186(6), 536–545. https://doi.org/10.1164/rccm.201204-0693OC
- Cui, L., Morris, A., Huang, L., Beck, J. M., Twigg, H. L., von Mutius, E., & Ghedin, E. (2014). The Microbiome and the Lung. Annals of the American Thoracic Society, 11(Supplement 4), S227–S232. https://doi.org/10.1513/AnnalsATS.201402-052PL
- de Steenhuijsen Piters, W. A. A., Huijskens, E. G. W., Wyllie, A. L., Biesbroek, G., van den Bergh, M. R., Veenhoven, R. H., Wang, X., Trzciński, K., Bonten, M. J., Rossen, J. W. A., Sanders, E. A. M., & Bogaert, D. (2016). Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. The ISME Journal, 10(1), 97–108. https://doi.org/10.1038/ismej.2015.99
- Denning, D. W., Pashley, C., Hartl, D., Wardlaw, A., Godet, C., del Giacco, S., Delhaes, L., & Sergejeva, S. (2014). Fungal allergy in asthma–state of the art and research needs. Clinical and Translational Allergy, 4(1), 14. https://doi.org/10.1186/2045-7022-4-14
- Dickson, R. P., & Huffnagle, G. B. (2015). The Lung Microbiome: New Principles for Respiratory Bacteriology in Health and Disease. PLOS Pathogens, 11(7), e1004923-. https://doi.org/10.1371/journal.ppat.1004923
- Dickson, R. P., Martinez, F. J., & Huffnagle, G. B. (2014). The role of the microbiome in exacerbations of chronic lung diseases. The Lancet, 384(9944), 691–702. https://doi.org/10.1016/S0140-6736(14)61136-3
- Dominguez-Bello, M. G., Costello, E. K., Contreras, M., Magris, M., Hidalgo, G., Fierer, N., & Knight, R. (2010). Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences, 107(26), 11971. https://doi.org/10.1073/pnas.1002601107
- Durack, J., Boushey, H. A., & Lynch, S. v. (2016). Airway Microbiota and the Implications of Dysbiosis in Asthma. Current Allergy and Asthma Reports, 16(8), 52. https://doi.org/10.1007/s11882-016-0631-8
- Fall, T., Lundholm, C., Örtqvist, A. K., Fall, K., Fang, F., Hedhammar, Å., Kämpe, O., Ingelsson, E., & Almqvist, C. (2015). Early Exposure to Dogs and Farm Animals and the Risk of Childhood Asthma. JAMA Pediatrics, 169(11), e153219–e153219. https://doi.org/10.1001/jamapediatrics.2015.3219
- File, T. M. (2003). Community-acquired pneumonia. The Lancet, 362(9400), 1991–2001. https://doi.org/10.1016/S0140-6736(03)15021-0
- Fujimura, K. E., Johnson, C. C., Ownby, D. R., Cox, M. J., Brodie, E. L., Havstad, S. L., Zoratti, E. M., Woodcroft, K. J., Bobbitt, K. R., Wegienka, G., Boushey, H. A., & Lynch, S. v. (2010). Man’s best friend? The effect of pet ownership on house dust microbial communities. The Journal of Allergy and Clinical Immunology, 126(2), 410-412.e4123. https://doi.org/10.1016/j.jaci.2010.05.042
- Fukata, M., & Arditi, M. (2013). The role of pattern recognition receptors in intestinal inflammation. Mucosal Immunology, 6(3), 451–463. https://doi.org/10.1038/mi.2013.13
- Hakansson, A. P., Orihuela, C. J., & Bogaert, D. (2018). Bacterial-Host Interactions: Physiology and Pathophysiology of Respiratory Infection. Physiological Reviews, 98(2), 781–811. https://doi.org/10.1152/physrev.00040.2016
- Hamilos, D. L. (2019). Biofilm Formations in Pediatric Respiratory Tract Infection. Current Infectious Disease Reports, 21(2), 6. https://doi.org/10.1007/s11908-019-0658-9
- Hanada, S., Pirzadeh, M., Carver, K. Y., & Deng, J. C. (2018). Respiratory Viral Infection-Induced Microbiome Alterations and Secondary Bacterial Pneumonia. Frontiers in Immunology, 9, 2640. https://www.frontiersin.org/article/10.3389/fimmu.2018.02640
- Hilty, M., Burke, C., Pedro, H., Cardenas, P., Bush, A., Bossley, C., Davies, J., Ervine, A., Poulter, L., Pachter, L., Moffatt, M. F., & Cookson, W. O. C. (2010). Disordered Microbial Communities in Asthmatic Airways. PLOS ONE, 5(1), e8578-. https://doi.org/10.1371/journal.pone.0008578
- Høiby, N., Ciofu, O., & Bjarnsholt, T. (2010). Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiology, 5(11), 1663–1674. https://doi.org/10.2217/fmb.10.125
- Huang, Y. J., & Boushey, H. A. (2015). The microbiome in asthma. Journal of Allergy and Clinical Immunology, 135(1), 25–30. https://doi.org/10.1016/j.jaci.2014.11.011
- Huang, Y. J., Sethi, S., Murphy, T., Nariya, S., Boushey, H. A., & Lynch, S. V. (2014). Airway microbiome dynamics in exacerbations of chronic obstructive pulmonary disease. Journal of clinical microbiology, 52(8), 2813–2823. https://doi.org/10.1128/JCM.00035-14
- Iwai, S., Huang, D., Fong, S., Jarlsberg, L. G., Worodria, W., Yoo, S., Cattamanchi, A., Davis, J. L., Kaswabuli, S., Segal, M., Huang, L., & Lynch, S. v. (2014). The Lung Microbiome of Ugandan HIV-Infected Pneumonia Patients Is Compositionally and Functionally Distinct from That of San Franciscan Patients. PLOS ONE, 9(4), e95726-. https://doi.org/10.1371/journal.pone.0095726
- Jain, S., Self, W. H., Wunderink, R. G., Fakhran, S., Balk, R., Bramley, A. M., Reed, C., Grijalva, C. G., Anderson, E. J., Courtney, D. M., Chappell, J. D., Qi, C., Hart, E. M., Carroll, F., Trabue, C., Donnelly, H. K., Williams, D. J., Zhu, Y., Arnold, S. R., … Finelli, L. (2015). Community-Acquired Pneumonia Requiring Hospitalization among U.S. Adults. New England Journal of Medicine, 373(5), 415–427. https://doi.org/10.1056/NEJMoa1500245
- Keogh, R. H., & Stanojevic, S. (2018). A guide to interpreting estimated median age of survival in cystic fibrosis patient registry reports. Journal of Cystic Fibrosis, 17(2), 213–217. https://doi.org/10.1016/j.jcf.2017.11.014
- Khalkhali, H. R., Oshnouei, S., Salarilak, S., Rahimi Rad, M., Karamyar, M., & Khashabi, J. (2014). Effects of antibiotic consumption on children 2-8 years of age developing asthma. Epidemiology and Health, 36, e2014006–e2014006. https://doi.org/10.4178/epih/e2014006
- Krone, C. L., Biesbroek, G., Trzciński, K., Sanders, E. A. M., & Bogaert, D. (2014). Respiratory microbiota dynamics following Streptococcus pneumoniae acquisition in young and elderly mice. Infection and Immunity, 82(4), 1725–1731. https://doi.org/10.1128/iai.01290-13
- Mallia, P., Message, S. D., Gielen, V., Contoli, M., Gray, K., Kebadze, T., Aniscenko, J., Laza-Stanca, V., Edwards, M. R., Slater, L., Papi, A., Stanciu, L. A., Kon, O. M., Johnson, M., & Johnston, S. L. (2011). Experimental Rhinovirus Infection as a Human Model of Chronic Obstructive Pulmonary Disease Exacerbation. American Journal of Respiratory and Critical Care Medicine, 183(6), 734–742. https://doi.org/10.1164/rccm.201006-0833OC
- Mika, M., Mack, I., Korten, I., Qi, W., Aebi, S., Frey, U., Latzin, P., & Hilty, M. (2015). Dynamics of the nasal microbiota in infancy: A prospective cohort study. Journal of Allergy and Clinical Immunology, 135(4), 905-912.e11. https://doi.org/10.1016/j.jaci.2014.12.1909
- Millares, L., Ferrari, R., Gallego, M., Garcia-Nuñez, M., Pérez-Brocal, V., Espasa, M., Pomares, X., Monton, C., Moya, A., & Monsó, E. (2014). Bronchial microbiome of severe COPD patients colonised by Pseudomonas aeruginosa. European Journal of Clinical Microbiology & Infectious Diseases, 33(7), 1101–1111. https://doi.org/10.1007/s10096-013-2044-0
- Nguyen, L. D. N., Deschaght, P., Merlin, S., Loywick, A., Audebert, C., van Daele, S., Viscogliosi, E., Vaneechoutte, M., & Delhaes, L. (2016). Effects of Propidium Monoazide (PMA) Treatment on Mycobiome and Bacteriome Analysis of Cystic Fibrosis Airways during Exacerbation. PLOS ONE, 11(12), e0168860-. https://doi.org/10.1371/journal.pone.0168860
- Nguyen, L. D. N., Viscogliosi, E., & Delhaes, L. (2015). The lung mycobiome: an emerging field of the human respiratory microbiome. Frontiers in Microbiology, 6, 89. https://www.frontiersin.org/article/10.3389/fmicb.2015.00089
- Ober, C., & Yao, T.-C. (2011). The genetics of asthma and allergic disease: a 21st century perspective. Immunological Reviews, 242(1), 10–30. https://doi.org/10.1111/j.1600-065X.2011.01029.x
- Orazi, G., & O’Toole, G. A. (2021). Pseudomonas aeruginosa Alters Staphylococcus aureus Sensitivity to Vancomycin in a Biofilm Model of Cystic Fibrosis Infection. MBio, 8(4), e00873-17. https://doi.org/10.1128/mBio.00873-17
- Ownby, D. R., Johnson, C. C., & Peterson, E. L. (2002). Exposure to Dogs and Cats in the First Year of Life and Risk of Allergic Sensitization at 6 to 7 Years of Age. JAMA, 288(8), 963–972. https://doi.org/10.1001/jama.288.8.963
- Pashley, C. H., Fairs, A., Free, R. C., & Wardlaw, A. J. (2012). DNA analysis of outdoor air reveals a high degree of fungal diversity, temporal variability, and genera not seen by spore morphology. Fungal Biology, 116(2), 214–224. https://doi.org/10.1016/j.funbio.2011.11.004
- Pettigrew, M. M., Tanner, W., & Harris, A. D. (2021). The Lung Microbiome and Pneumonia. The Journal of Infectious Diseases, 223(Supplement_3), S241–S245. https://doi.org/10.1093/infdis/jiaa702
- Popgeorgiev, N., Temmam, S., Raoult, D., & Desnues, C. (2013). Describing the Silent Human Virome with an Emphasis on Giant Viruses. Intervirology, 56(6), 395–412. https://doi.org/10.1159/000354561
- Prescott, S. L. (2013). Early-life environmental determinants of allergic diseases and the wider pandemic of inflammatory noncommunicable diseases. Journal of Allergy and Clinical Immunology, 131(1), 23–30. https://doi.org/https://doi.org/10.1016/j.jaci.2012.11.019
- Price, K. E., Hampton, T. H., Gifford, A. H., Dolben, E. L., Hogan, D. A., Morrison, H. G., Sogin, M. L., & O’Toole, G. A. (2013). Unique microbial communities persist in individual cystic fibrosis patients throughout a clinical exacerbation. Microbiome, 1(1), 27. https://doi.org/10.1186/2049-2618-1-27
- Ramos-Sevillano, E., Wade, W. G., Mann, A., Gilbert, A., Lambkin-Williams, R., Killingley, B., Nguyen-Van-Tam, J. S., & Tang, C. M. (2019). The Effect of Influenza Virus on the Human Oropharyngeal Microbiome. Clinical Infectious Diseases, 68(12), 1993–2002. https://doi.org/10.1093/cid/ciy821
- Rohde, G., Wiethege, A., Borg, I., Kauth, M., Bauer, T. T., Gillissen, A., Bufe, A., & Schultze-Werninghaus, G. (2003). Respiratory viruses in exacerbations of chronic obstructive pulmonary disease requiring hospitalisation: a case-control study. Thorax, 58(1), 37. https://doi.org/10.1136/thorax.58.1.37
- SEEMUNGAL, T., HARPER-OWEN, R., BHOWMIK, A., MORIC, I., SANDERSON, G., MESSAGE, S., MacCALLUM, P., MEADE, T. W., JEFFRIES, D. J., JOHNSTON, S. L., & WEDZICHA, J. A. (2001). Respiratory Viruses, Symptoms, and Inflammatory Markers in Acute Exacerbations and Stable Chronic Obstructive Pulmonary Disease. American Journal of Respiratory and Critical Care Medicine, 164(9), 1618–1623. https://doi.org/10.1164/ajrccm.164.9.2105011
- Shilts, M. H., Rosas-Salazar, C., Tovchigrechko, A., Larkin, E. K., Torralba, M., Akopov, A., Halpin, R., Peebles, R. S., Moore, M. L., Anderson, L. J., Nelson, K. E., Hartert, T. v, & Das, S. R. (2016). Minimally Invasive Sampling Method Identifies Differences in Taxonomic Richness of Nasal Microbiomes in Young Infants Associated with Mode of Delivery. Microbial Ecology, 71(1), 233–242. https://doi.org/10.1007/s00248-015-0663-y
- Stearns, J. C., Davidson, C. J., McKeon, S., Whelan, F. J., Fontes, M. E., Schryvers, A. B., Bowdish, D. M. E., Kellner, J. D., & Surette, M. G. (2015). Culture and molecular-based profiles show shifts in bacterial communities of the upper respiratory tract that occur with age. The ISME Journal, 9(5), 1246–1259. https://doi.org/10.1038/ismej.2014.250
- Sze, M. A., Dimitriu, P. A., Suzuki, M., McDonough, J. E., Campbell, J. D., Brothers, J. F., Erb-Downward, J. R., Huffnagle, G. B., Hayashi, S., Elliott, W. M., Cooper, J., Sin, D. D., Lenburg, M. E., Spira, A., Mohn, W. W., & Hogg, J. C. (2015). Host Response to the Lung Microbiome in Chronic Obstructive Pulmonary Disease. American Journal of Respiratory and Critical Care Medicine, 192(4), 438–445. https://doi.org/10.1164/rccm.201502-0223OC
- Teo, S. M., Mok, D., Pham, K., Kusel, M., Serralha, M., Troy, N., Holt, B. J., Hales, B. J., Walker, M. L., Hollams, E., Bochkov, Y. A., Grindle, K., Johnston, S. L., Gern, J. E., Sly, P. D., Holt, P. G., Holt, K. E., & Inouye, M. (2015). The Infant Nasopharyngeal Microbiome Impacts Severity of Lower Respiratory Infection and Risk of Asthma Development. Cell Host & Microbe, 17(5), 704–715. https://doi.org/https://doi.org/10.1016/j.chom.2015.03.008
- Teo, S. M., Mok, D., Pham, K., Kusel, M., Serralha, M., Troy, N., Holt, B. J., Hales, B. J., Walker, M. L., Hollams, E., Bochkov, Y. A., Grindle, K., Johnston, S. L., Gern, J. E., Sly, P. D., Holt, P. G., Holt, K. E., & Inouye, M. (2015). The Infant Nasopharyngeal Microbiome Impacts Severity of Lower Respiratory Infection and Risk of Asthma Development. Cell Host & Microbe, 17(5), 704–715. https://doi.org/10.1016/j.chom.2015.03.008
- Underhill, D. M., & Iliev, I. D. (2014). The mycobiota: interactions between commensal fungi and the host immune system. Nature Reviews Immunology, 14(6), 405–416. https://doi.org/10.1038/nri3684
- Unger, S. A., & Bogaert, D. (2017). The respiratory microbiome and respiratory infections. Journal of Infection, 74, S84–S88. https://doi.org/10.1016/S0163-4453(17)30196-2
- Urb, M., Snarr, B. D., Wojewodka, G., Lehoux, M., Lee, M. J., Ralph, B., Divangahi, M., King, I. L., McGovern, T. K., Martin, J. G., Fraser, R., Radzioch, D., & Sheppard, D. C. (2015). Evolution of the Immune Response to Chronic Airway Colonization with Aspergillus fumigatus Hyphae. Infection and immunity, 83(9), 3590–3600. https://doi.org/10.1128/IAI.00359-15
- Valley, T. S., Sjoding, M. W., Ryan, A. M., Iwashyna, T. J., & Cooke, C. R. (2015). Association of Intensive Care Unit Admission With Mortality Among Older Patients With Pneumonia. JAMA, 314(12), 1272–1279. https://doi.org/10.1001/jama.2015.11068
- van Woerden, H. C., Gregory, C., Brown, R., Marchesi, J. R., Hoogendoorn, B., & Matthews, I. P. (2013). Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: a community based case control study. BMC Infectious Diseases, 13(1), 69. https://doi.org/10.1186/1471-2334-13-69
- Wang, Z., Bafadhel, M., Haldar, K., Spivak, A., Mayhew, D., Miller, B. E., Tal-Singer, R., Johnston, S. L., Ramsheh, M. Y., Barer, M. R., Brightling, C. E., & Brown, J. R. (2016). Lung microbiome dynamics in COPD exacerbations. European Respiratory Journal, 47(4), 1082. https://doi.org/10.1183/13993003.01406-2015
- Wardlaw, T., Salama, P., Johansson, E. W., & Mason, E. (2006). Pneumonia: the leading killer of children. The Lancet, 368(9541), 1048–1050. https://doi.org/10.1016/S0140-6736(06)69334-3
- Watson, R. L., de Koff, E. M., & Bogaert, D. (2019). Characterising the respiratory microbiome. European Respiratory Journal, 53(2), 1801711. https://doi.org/10.1183/13993003.01711-2018
- Willner, D., Furlan, M., Haynes, M., Schmieder, R., Angly, F. E., Silva, J., Tammadoni, S., Nosrat, B., Conrad, D., & Rohwer, F. (2009). Metagenomic Analysis of Respiratory Tract DNA Viral Communities in Cystic Fibrosis and Non-Cystic Fibrosis Individuals. PLOS ONE, 4(10), e7370-. https://doi.org/10.1371/journal.pone.0007370
- Zaura, E., Nicu, E. A., Krom, B. P., & Keijser, B. J. F. (2014). Acquiring and maintaining a normal oral microbiome: current perspective. Frontiers in Cellular and Infection Microbiology, 4, 85. https://www.frontiersin.org/article/10.3389/fcimb.2014.00085
- Zhao, J., Schloss, P. D., Kalikin, L. M., Carmody, L. A., Foster, B. K., Petrosino, J. F., Cavalcoli, J. D., VanDevanter, D. R., Murray, S., Li, J. Z., Young, V. B., & LiPuma, J. J. (2012). Decade-long bacterial community dynamics in cystic fibrosis airways. Proceedings of the National Academy of Sciences, 109(15), 5809. https://doi.org/10.1073/pnas.1120577109
- Zhou, F., Yu, T., Du, R., Fan, G., Liu, Y., Liu, Z., Xiang, J., Wang, Y., Song, B., Gu, X., Guan, L., Wei, Y., Li, H., Wu, X., Xu, J., Tu, S., Zhang, Y., Chen, H., & Cao, B. (2020). Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The Lancet, 395(10229), 1054–1062. https://doi.org/10.1016/S0140-6736(20)30566-3

