14 Plant Microbiomes
Plant Microbiomes
Plant health is influenced by a variety of environmental factors, and though the soil content (including the microbial community present) is a major element, the distinct plant microbiome serves an important role. Similar to organ systems in the human body (e.g. skin, gut, etc.), the components of various plants (e.g. internal tissues, leaves, roots, etc.) can have unique microbial communities that contribute to its respective and overall vitality. A better understanding of the corresponding roles of plant microbiomes can be combined in a synergistic effort with other environmental microbiomes to maintain and promote sustainable agricultural practices, ecosystem biodiversity, and research model studies.
Plant microbiome–an account of the factors that shape community composition and diversity
Article by Dastogeer et al., 2020 licensed under the terms of the Creative Commons Attribution License (CC BY).
Abstract
Plants live in association with diverse microorganisms, collectively called the microbiome. These microbes live either inside (endosphere) or outside (episphere) of plant tissues. Microbes play important roles in the ecology and physiology of plants. Significant progress has been made in revealing structure and dynamics of plant microbiome in the last few years. Various factors related to host, microbes as well as environment influence the community composition and diversity of plant microbiome. This review aimed to provide a general account of the factors (host, microbe and environment) that drive the microbial community composition in plant. First, we gave an overview of the aboveground and belowground plant microbiomes. Next, we discussed which host factors are involved in variation in plants followed by importance of microbe-microbe interactions and the elements of environment that influence composition and community structuring of plant microbiomes.
1. Introduction
A diverse kind of microorganisms associated with a higher organism (human, animal, plants etc.) is together defined as its microbiome. All higher organisms examined to date, including plants, insects, fish, rats, apes, and humans, harbor microbiomes [1,2]. Research on the human microbiome has progressed very quickly. Recently, researchers have also paid much attention to elucidating the composition and functions of plant and soil microbiomes. It is now believed that plants are not separate entities, but rather they live in association with a large variety of microbes. These microbes live either inside (endosphere) or outside (episphere) of plant tissues. Among these microorganisms, bacteria and fungi are predominant. About a few thousand bacterial and fungal taxa have been reported from plant tissues [3,4]. They play important roles such as increased nutrient availability, uptake by plants and increased plant stress tolerance. Thus, plant fitness (growth and survival) is the result of physical and physiological functions of the plant per se as well as the associated microbiome, which together are known as a plant holobiont [5].
The study of the association of plants with microorganisms precedes that of the animal and human microbiomes, notably the roles of microbes in nitrogen and phosphorus uptake. The most notable examples are plant root-arbuscular mycorrhizal (AM) and legume-rhizobial symbioses, both of which greatly influence the ability of roots to uptake various nutrients from the soil. Some of these microbes cannot survive in the absence of the plant host (the ‘obligate symbionts’ including viruses, some bacteria and fungi), which provides space, oxygen, proteins, and carbohydrates to the microorganisms. The association of AM fungi with plants has been known since 1842, and over 80 % of land plants are found associated with them [6]. It is thought that AM fungi helped in the domestication of plants [7]. Traditionally, culturable microbes have been used for plant-microbe interaction studies with the enormous unculturable microbes remain uninvestigated and consequently, our knowledge of the roles of these unculturable microbes remains largely unknown.
Unraveling the types and outcomes of plant-microbe interactions has received considerable interest among ecologists, evolutionary biologists, plant biologists, and agronomists [4,8,9]. Recent developments in meta-omics and the establishment of large collections of microorganisms have dramatically increased our knowledge of the plant microbiome composition and diversity. The sequencing of marker genes of entire microbial communities, referred to as metagenomics, sheds light on the phylogenetic diversity of the microbiomes of plants. It also adds to the knowledge of the major biotic and abiotic factors responsible for shaping plant microbiome community assemblages [8].
However, our understanding on the roles of microbiomes, with respect to their impact on plant ecology and physiology, is still far from complete, and we are at the beginning of knowing their functions [10]. The outcome of this improved knowledge will have significant bearings on a variety of experiments and applications, such as development of biofertilizer and biopesticides for sustainable agricultural production with less reliance on agrochemicals, while augmenting yield and nutritional value [11].
In this review, we will present an account of recent studies and prospects of studying the plant microbiome. Firstly, we will present an overview of aboveground and belowground plant microbiomes. Next, we will discuss the factors driving the composition and community structuring of plant microbiomes. We will not address plant pathogenic microbes, although inclusion of this fraction of microbiome would make sense from a broader and more holistic viewpoint.
2. Rhizosphere microbiome
The rhizosphere comprises the 1–10 mm zone of soil immediately surrounding the roots that is under the influence of the plant through its deposition of root exudates, mucilage and dead plant cells [12]. A diverse array of organisms specialize in living in the rhizosphere, including bacteria, fungi, oomycetes, nematodes, algae, protozoa, viruses, and archaea [13]. The most frequently studied beneficial rhizosphere organisms are mycorrhizae, rhizobium bacteria, plant growth promoting rhizobacteria (PGPR), and biocontrol microbes. Gans, Wolinsky [14] projected that one gram of soil could harbor more than a million distinct bacterial genomes. İnceoğlu, Al-Soud [15] reported 55,121 OTUs (operational taxonomic unites) from the potato rhizosphere. Among the prokaryotes in the rhizosphere, the most frequent bacteria are within the Acidobacteria, Proteobacteria, Planctomycetes, Actinobacteria, Bacteroidetes, and Firmicutes [3,16]. In some studies, no significant differences were reported in the microbial community composition between the bulk soil (soil not attached to the plant root) and rhizosphere soil [17,18]. Certain bacterial groups (e. g. Actinobacteria, Xanthomonadaceae) are less abundant in the rhizosphere than in nearby bulk soil [3].
Mycorrhizal fungi are abundant members of the rhizosphere community, and have been found in over 200,000 plant species, and are estimated to associate with over 80 % of all plants [19]. These mycorrhizae–root associations play profound roles in land ecosystems by regulating nutrient and carbon cycles. Mycorrhizae are integral to plant health because they provide up to 80 % of N and P requirements. In return, the fungi obtain carbohydrates and lipids from host plants [20]. Recent studies of arbuscular mycorrhizal fungi using sequencing technologies show greater between-species and within-species diversity than previously known [21].
3. Phyllosphere microbiome
The aerial surface of a plant (stem, leaf, flower, fruit) is called the phyllosphere and is considered comparatively nutrient poor when compared to the rhizosphere and endosphere. The environment in the phyllosphere is more dynamic than the rhizosphere and endosphere environments. Microbial colonizers are subjected to diurnal and seasonal fluctuations of heat, moisture, and radiation. In addition, these environmental elements affect plant physiology (such as photosynthesis, respiration, water uptake etc.) and indirectly influence microbiome composition. Rain and wind also cause temporal variation to the phyllosphere microbiome [22]. Overall, there remains high species richness in phyllosphere communities. Fungal communities are highly variable in the phyllosphere of temperate regions and are more diverse than in tropical regions [23]. There can be up to 107 microbes per cm2 present on leaf surfaces of plants, and thus the bacterial population of the phyllosphere on a global scale is estimated to be 1026 cells [24]. The population size of the fungal phyllosphere is likely to be smaller [25]. Phyllosphere microbes from different plants appear to be somewhat similar at high levels of taxa, but at the lower levels taxa there remain significant differences. This indicates that microorganisms may need finely tuned metabolic adjustment to survive in phyllosphere environment [24]. Proteobacteria seems to be the dominant colonizers, with Bacteroidetes and Actinobacteria also predominant in phyllospheres [26]. Although there are similarities between the rhizosphere and soil microbial communities, very low similarity has been reported between phyllosphere communities and those in open air [27].
4. Endosphere microbiome
Some microorganisms, such as endophytes, penetrate and occupy the plant internal tissues, forming the endospheric microbiome (Fig. 1). The AM and other endophytic fungi are the dominant colonizers of the endosphere [28]. Bacteria, and to some degree Archaea, are important members of endosphere communities. Some of these endophytic microbes interact with their host and provide obvious benefits to plants [[29], [30], [31]]. Unlike the rhizosphere and the rhizoplane, the endospheres harbor highly specific microbial communities. The root endophytic community can be very distinct from that of the adjacent soil community. In general, diversity of the endophytic community is lower than the diversity of the microbial community outside the plant [18]. The identity and diversity of the endophytic microbiome of above-and below-ground tissues may also differ within the plant [28].
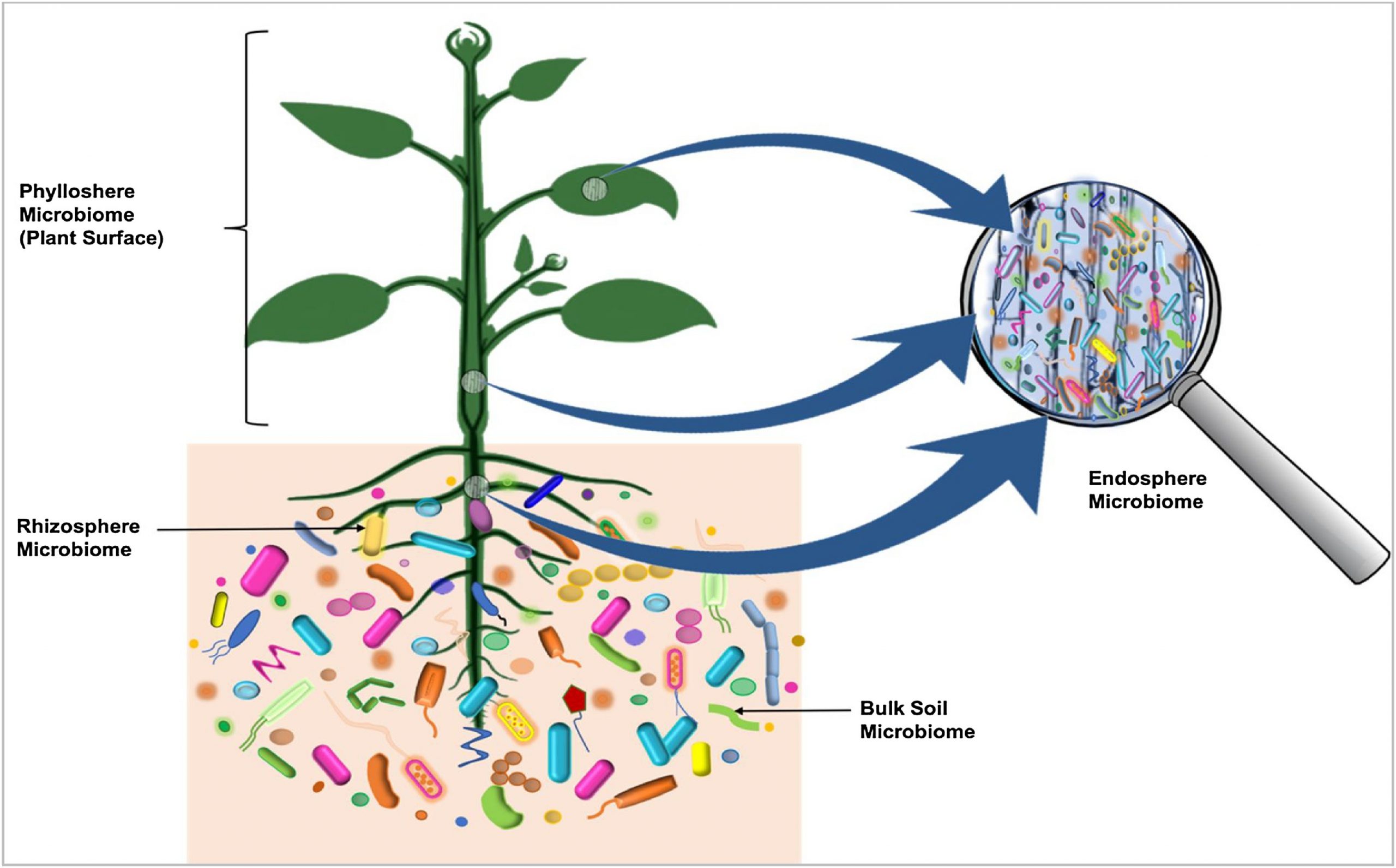
5. Drivers of plant microbiome composition
Plant microbiome structure is influenced by complex interactions between hosts, microbes, and associated environmental factors such as climate, soil, cultivation practices etc. (Fig. 2). Below, we provide an assessment of current knowledge of these factors, providing insight to plant-microbe interactions in a broader-sense.
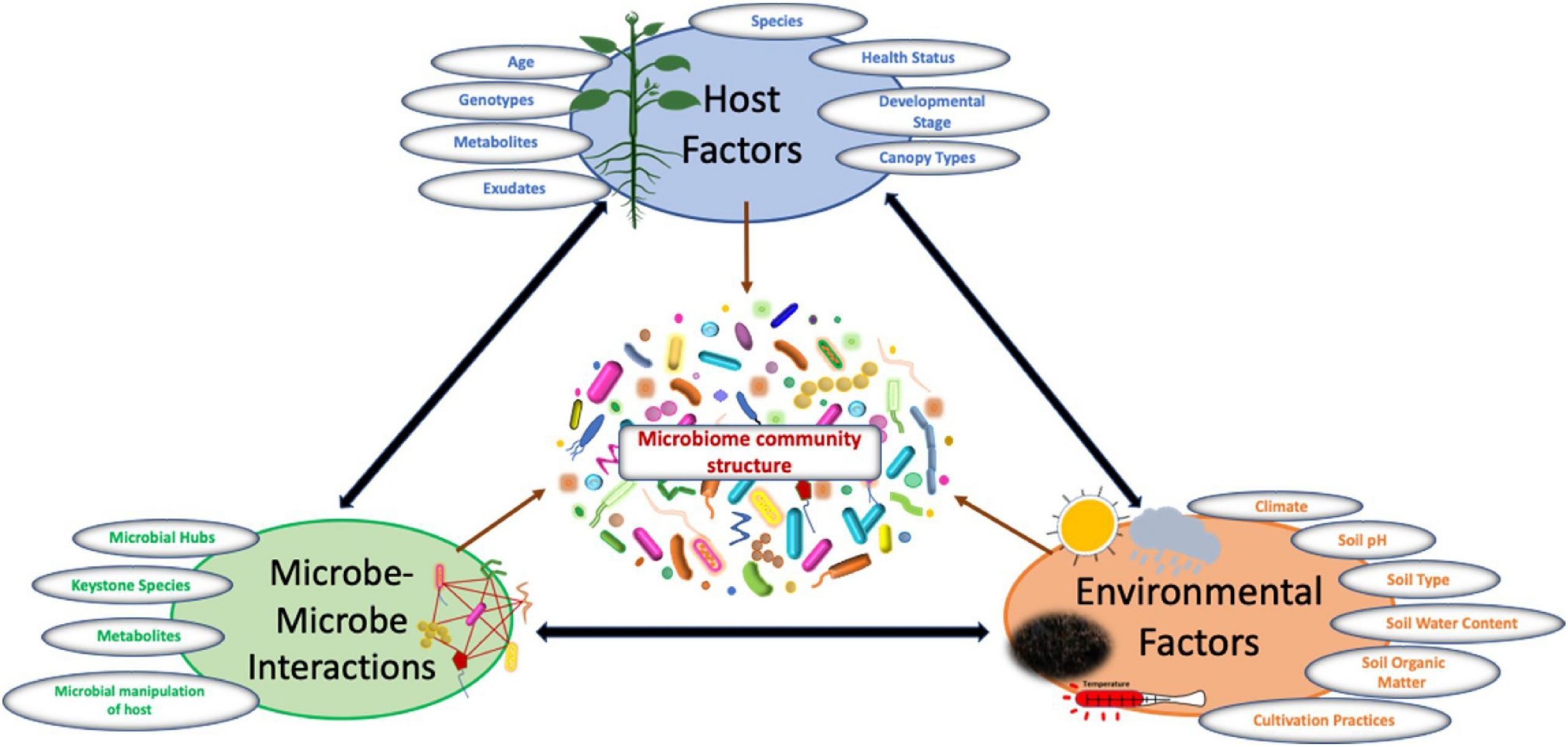
6. Host factors that influence plant microbiome community composition
6.1. Plant species
The identity of the host plant has a significant influence on the identity of its microbiome. Different plant species growing adjacent to one another can harbor distinct microbiomes. A comparative survey of root microbiomes in maize, sorghum, and wheat showed different community composition among these plants [32]. Samad et al. [33] investigated the microbiome compositions of roots and rhizospheres using 16S rRNA gene from grapevines and some weed species growing in the same field, and their findings suggested that these species hosted significantly different microbiomes in the roots and rhizosphere, with the more pronounced difference in the root communities. Plants that are distantly-related phylogenetically show greater variation in associated microbiome compositions, suggesting a role of plant phylogeny in structuring root microbiomes [32]. Plant species also influences the identity and diversity of endophytic communities. Manter et al. [34] reported differences in endophytic community composition in potato and Eucalyptus plants. The most abundant bacterial root endophytes in potato were rare or absent in Eucalyptus and vice-versa [34] suggesting that the host plant selects its endophytic microbes. Analysis of endophytic fungi of three native Australian Nicotiana species revealed that they are host-specific but not plant organ- or host location-specific [28]. In addition to the rhizospheric and endophytic microbiomes, phyllosphere community composition also depends on plant identity [24]. Kembel et al. [35] showed that the leaf microbiome community is highly correlated with plant evolutionary relatedness similar to the endospheric microbiome.
The effects of host plant species in recruiting microbes from the surrounding environment indicate that plants have evolved traits that govern root microbiome assemblages [36]. For example, endosphere, rhizosphere community composition are correlated with host taxonomy [36]. Xiao et al. [37] found that the rhizosphere and root microbiomes are mostly influenced by soil type, and the nodule and root endophytes are influenced by plant species. Differential microbiome assembly in different plant species is attributed to variation in plant resource consumption [36]. Plant traits such as leaf permeability, wettability and topography and physicochemical properties, cuticle chemistry, root exudates, antibiotic production, and inherent plant immunity to invasion by microbes may also to play a role.
6.2. Plant genotypes
Evidence suggests a difference in microbe community composition between genotypes of a particular species [17,26,38]. Genetics of the host is one of the factors that shape the plant-microbiome structure. For example, OTUs in three different potato varieties were cultivar-specific [36]. Similarly, cultivar-dependent effects have been reported for the bacterial communities in young potato rhizospheres [15]. Peiffer et al. [39] demonstrated that OTU richness and β-diversity are influenced by plant genotypes in maize. Bulgarelli et al. [38] reported that genotype contributed to about 6% of the variation of the microbiome composition in the rhizosphere region. A larger influence of host genotype on community composition has been reported [40]. Genotype-dependent microbiome community structuring has been reported for sweet potato, wheat, pea, and oat [9,41]. Bacteria such as Acinetobacter, Chryseobacterium, Pseudomonas, Sphingobium, and Stenotrophomonas were more abundant in low-starch cultivars than those having high-starch contents [41]. The rhizosphere communities of different genetic clones of wild-type and transgenic lines have been reported to be distinct in Populus [42]. Within-species genetic variability can influence microbiome composition in leaf tissues [43]. Wagner et al. [44] conducted an extensive field experiment to unravel drivers of community composition of bacteria associated with leaves and roots of Boechera stricta. Their findings suggested that the host genotype influences leaf community, but the root microbiome was variable at different collection sites.
Agler et al. [45] proposed that host genotype influences on keystone microbes, which then pass these effects onto the total microbiome by changing microbe–microbe interactions and altering plant fitness [3]. The host specificity of plant microbiome could also be attributed to the nutrient preferences of plants [46]. However, whether the observed host influences are heritable, how strong the effect is, and whether these associations will be actionable for plant breeding is yet to be established.
6.3. Plant organ
Different plant tissues host distinct microbiome communities. Edwards et al. [47] demonstrated that each surface and internal tissue of plants may harbor distinct microbial communities and that the role of tissue-type was greater than host type and the microbiome of the soil. This may be because the adaptation strategies of various tissues may affect the microbes in colonizing them for community composition. For instance, surface tissues are exposed to constant fluctuations of weather and have relatively poor nutritional status compared to the root or internal tissues. Therefore, microbes colonizing the leaf surface need to be adapted in these conditions [25]. Other studies found very little or no effect of plant organ in community composition of fungi and bacteria [28].
Study with agaves determined that prokaryotes are largely influenced by the plant compartment, whereas the rhizosphere, the phyllosphere, and endosphere communities are clearly different from each other and from adjacent soils [48]. Many studies have reported that fungal communities show a different pattern, where the biogeography of host were the major influencing factor [48]. This may be explained by the dispersal limitation in fungi [49], because fungi are eukaryotes like plants and animals, but bacteria are different in this respect.
6.4. Plant age and developmental stage
In the cases of interactions between plants and pathogens, ontogenic resistance (age-related resistance) is widely reported and correlated with plant developmental stage [50]. Symbiosis research has also indicated that plant age and developmental stage are important factors affecting microbial communities [51]. Analysis of the bacterial rhizosphere community of Arabidopsis revealed that the seedling stage selects distinct microbiomes at developmental time points. Plants produce mixtures of compounds and specific phytochemicals in the root exudates. Some of these chemicals are indeed distinct at plant developmental stages and appear to shape microbiome community assemblages [52]. The effect of the plant age on microbiome using DGGE fingerprint analysis revealed that it significantly influences bacterial community composition of all groups investigated for all three sweet potato cultivars [41]. The effect of plant age on the composition of bacterial microbiome in the rhizosphere has been demonstrated in potato, maize and soybean [15,53,54]. Age-related microbiome differentiation may be associated with root growth, physiology, root architecture, root morphology, root exudate, and its composition [51,52]. However, further research is needed on the identity and effect of root exudates at different plant developmental stages to determine how plants communicate in the rhizosphere. This knowledge might offer a basis for augmenting agricultural crops by the application of rhizosphere microbes.
6.5. Plant canopy type
Plant canopy type also influences microbiome community composition. For example, bacterial communities in sugar maple leaf samples are correlated with canopy composition [55]. The microbial migration through rain runoff may be an important factor for variation in microbial colonization in different canopy types. Canopy structure influences the composition of endophytic community but not the rhizospheric community, indicating less effect of rain runoff, and there may be other mechanisms such as soil factors or some unknown factors involved in this variation [55].
6.6. Plant immunity and signaling
Plant health status may influence microbiome composition. Plants employ two layers of defense against pathogens: pattern-triggered immunity (PTI), which is triggered by conserved molecular structures such as microbe/pathogen-associated molecular patterns (MAMPs/PAMPs), and damage-associated molecular patterns, which are recognized by plasma membrane-localized pattern recognition receptors [56]. It is unknown whether plants recognize non-pathogenic microbes in the similar way as they recognize pathogens and modulate their response. When plants are challenged with herbivory or pathogens they release hormones and exogenous volatiles that alter the composition of root exudates (for review, see [57]), and these in turn modify the microbiome community. Aphid infestation and pathogenic microbial infection increaseed populations of the non-pathogenic rhizobacterium Bacillus subtilis in the sweet pepper rhizosphere [58]. In Arabidopsis thaliana plants infected by Pseudomonas syringae, the expression of root malate transporter is altered, indicating a change in secretion of malic acid that increased the number of the beneficial rhizobacterium Bacillus subtilis [59,60]. Cucumber roots infected by Fusarium oxysporum f. sp. cucumerinum exhibited augmented secretion of fumaric and citric acid, which led to the formation of biofilms (aggregates of living bacteria in a slimy extracellular polymeric substances) of Bacillus amyloliquefaciens. Most investigations, however, have focused on one-to-one interactions (plant-microbe), although in reality, plants are subjected to attack by numerous microbial pathogens and insect pests. Therefore, it would be interesting to see how multiple herbivory and/or pathogens modify the community composition of plant microbiomes. Recent studies have reported a change in rhizosphere microbiome community composition as influenced by specific compounds such as sugars, sugar alcohols, or mixtures of various chemical compounds in root exudates [52]. Plants use various strategies in response to pathogenic infection and insect attack. One of them is activation of defense responses in roots, which may influence microbiome composition in the rhizosphere and roots [61]. When aphids feed on foliage both SAR (Systemic Acquired Resistance) and ISR (Induced Systemic Resistance) signaling are activated throughout the plants, which elicits sweet pepper plants to attract B. subtilis in the rhizosphere [58]. Again, increased JA signaling in plant either by injury or exogenously in Medicago truncatula caused higher colonization of beneficial mycorrhizae [62]. Different root and phyllosphere endophyte microbial communities have been reported when altered SA signaling was induced [63]. Plants lacking in jasmonate-mediated defense have shown more diverse epiphytic colonization [64]. It is evident from these studies that the role of plant defense systems on the microbial composition are inconstant, and that SAR is an important factor in regulating some bacterial community composition. Chemical signals released by plants for example, flavonoids, activate varied responses in plant rhizosphere microbiomes [65]. Branching in mycorrhizal hyphae is affected by strigolactones, which enhance and promote seed germination by parasitic plants [66].
6.7. Plant derived compounds
A diverse array of antimicrobial compounds are produced in plants [67]. Some of them, such as different alkaloids, phenolics, and terpenoids, are common in plants. Some are specific to particular groups [68], for instance, Brassicales produce glucosinolates. It was found that transgenic Arabidopsis producing an exogenous glucosinolate had different microbiomes in the rhizosphere and root tissues [69]. Voges et al. 2018 reported a significant role of plant derived coumarins in structuring the rhizosphere community. They suggested that iron-mobilizing coumarins are involved in redox reactions that can mobilize ferric iron and generate reactive oxygen species (ROS) with detrimental effects on microbial proliferation and thus selectively inhibit certain microbial growth while allowing proliferation other more beneficial partners. Triterpenoid saponins, which are known as avenacins, are found in oat (Avena strigosa), and have antifungal properties [70]. Oat plants lacking avenacin production attracted different culturable fungi in roots [71] and were more vulnerable to pathogenic infections than wild-type oat [72]. Interestingly, however, a recent comprehensive analysis of the rhizosphere community of these two genotypes reported little difference between the fungal communities. The effect of avenacins on the Amoebozoa and Alveolata was profound but has not been reported for bacterial communities [73]. This revealed that a small difference in plant genotype might exert multifaceted and unpredicted effects on the plant microbiome composition and diversity. These plants derived compound may affect microbiome assembly in different ways. For example, root exudates may be specific for host plant and can modulate rhizosphere community as well as selects specific root microbiome and thus contributing host specific plant microbiome. Also, antimicrobial compound may selectively enhance microbial growth by restricting certain microbes which is a kind of ‘balance’ in mutualism.
7. Microbial factors in shaping plant microbiome structure
Microorganisms play important roles in shaping microbial community structures in plants. However, our knowledge on how microorganisms influence microbiome structuring is limited.
7.1. Microbial manipulation of hosts
Microorganisms can affect host plants, for example host root exudations, which in turn affect the permeability of roots and root metabolism. Some microbes in soil where the plant if growing can also absorb certain compounds in root exudates and excrete other compounds. Soil microbes can produce compounds that affect plant signaling and metabolism, which lead to production of microbe-derived compounds in plants. Some microbes produce antibiotics (e.g., penicillin and polymyxin) which increase the exudation of organic materials, altered cell permeability, and increased leakage [74] and results in a variable microbiome assembly.
7.2. Microbe-microbe interaction
The extent to which microbe–microbe interactions can play roles in the microbiome composition is not well understood. The outcome of microbe-microbe interactions could be explained as cooperation, parasitism, and competition. In cooperation, at least one species benefits, while others are not harmed. When both species benefit, the term mutualism is used, whereas, when one partner benefits while the other is not affected, the term commensalism is used. In contrast, parasitism and competition are harmful for at least one species [75].
We know from recent studies that microbial communities harbor highly connected taxa called keystone taxa [76]. These taxa independently or in a group show a substantial effect on microbiome composition and functions regardless of their spatial and temporal dynamics. They play a unique and vital role on microbiomes, and their absence could cause a significant alterations in microbiome composition and functioning [76]. They use various strategies to impact on host microbiome. For instance, they might cause changes in intermediate or effector groups which in turn regulate microbiome community composition and functioning [77]. Production of a secondary metabolite (2,4-diacetyl phloroglucinol) was reported for some strains of Pseudomonas fluorescens that suppress Gaeumannomyces graminis var. tritici, responsible for wheat take-all [78]. Similarly, many fungi (e.g. Trichoderma and bacteria (e.g. Bacillus) produce various secondary metabolites that suppress microbial growth [79,80]. The keystone taxa may produce bacteriocins to shift microbiota structure selectively. Again, by synergism, keystone taxa may alter the abundance of their partners, and influence community structure and performance. For example, certain species of Burkholderia are symbiotic with arbuscular mycorrhizae and may change abundance and community composition of AM fungi, thereby influencing plant community richness, diversity, and production [19]. Agler et al. [45] studied the roles of microorganisms (bacteria, fungi and oomycetes) in the community composition of phyllosphere microbiomes of Arabidopsis thaliana using a systems biology approach. They described an interkingdom interactions network with a profound influence on community structure. They identified a few taxa, termed “microbial hubs’, which are highly interlinked and have a significant impact on communities. They used two “hub” microbes (Albugo, an oomycete pathogen and Dioszegia, a basidiomycete yeast) in detail. Albugo strongly affected epiphytic and endophytic bacterial colonization. Many symbiotic microbes (including pathogens) produce effector proteins to suppress, activate, or alter host defense mechanisms [81], and some can entirely reprogram the host metabolism [82]. These host adjustments can lead to alterations of microbiome composition because some microorganisms and not others can benefit from altered conditions. Actually, the niches of some microorganisms is dependent on others. For instance, primary colonizers can aid subsequent colonizers against hazardous abiotic factors [83] or can enhance the competitive ability of following colonizers by producing secondary metabolic compounds [84]. There can be direct microbe-microbe interactions, such as the hyper-parasitism (parasite of parasite) of primary colonizers [85] and opportunists that utilize host’s compromised plant defenses to colonize them [86]. Such phenomena point out why some colonizers can affect the development of microbes on the host even if they may be distantly related [84] and highlight a crucial functions of such interactions in shaping microbiome composition and structure.
One of the major strategies by which PGPR augments plant growth is by its influence on rhizosphere microbes. For example, Pseudomonas sp. DSMZ 13134 alters the composition of dominant bacteria in barley roots [87]. In some cases, however, the effects of PGPRs on resident microbiome may not be prominent. For example, no substantial changes in rhizosphere community were noticed after the application of Bacillus amyloliquefaciens FZB42 [88]. Supporting results have also been reported in soybean with the application of B. amylolique faciens BNM122 [89]. The recent investigation on the effect of B. amyloli quefaciens on the lettuce microbiome using 454-amplicon sequencing revealed no or only transient and minor effects in the rhizosphere zone [90]. Interestingly, a decrease in the bacterial number was reported. In the field only 55 % of the inoculated bacterial DNA could be traced after a month [91]. The effects of Bacillus subtilis strain PTS-394 on the rhizosphere microbiome has been examined by metagenomic profiling. Similar to the results above for B. amyloliquefaciens FZB42 [90,91], only a minor effect on the composition was reported. However, up until now, the impact of Bacillus PGPR on other plant microbiota, such as fungi, has not been investigated, and investigation on this could reveal a more general effect of inoculated bacteria on resident microbiota and thus on host physiology and ecology.
8. Environmental factors as drivers of plant microbiome assembly
Environmental elements, such as soils, climatic conditions, geography, farming activities, and plant domestication, could result in the differences of plant associated microbial community composition [92]. A change in an environmental component results in plant phenotypic changes (e.g., [93], which consequently also change the assemblage of distinct microbiomes harboring plant compartments [94]. Plants grown under controlled conditions provide specific environments for microorganisms. For instance, when lettuce was grown under a glasshouse, a distinct bacterial signature was found from those grown in open fields [95]. Whitaker, Reynolds et al. [96] reported the community composition of endophytic fungi local environment (i.e., site), but not by host ecotype, pointing that environmental factors are major drivers of the endophytic mycobiome of switchgrass.
8.1. Soil
Plants recruit root microbes mainly from the soils where they grow. Various soil factors viz. soil types, soil pH, and the C/N ratio, as well as available P and K are frequently reported to be the determinants of root microbial community composition by affecting plant growth and immunity [8,9]. Innumerable studies using high throughput sequencing have proved that soil type is a major factor for root microbiome structure, which is evident from the differential initial microbial inocula present in different soil types [18,47]. Dombrowski et al. [97] collected the arctic-alpine Arabis alpine samples from the native location as well as from those grown under controlled conditions and investigated the root microbiome by 16S rRNA amplicon sequencing analysis. They reported that soil type and length of time the plants remained in the soil are the most important drivers, causing variation of up to 15 % of root microbiota. In addition, in the same soil, the root microbiome of perennial A. alpina was similar to A. thaliana and Cardamine hirsuta, the annual relatives of A. alpina. The root microbiome communities are strongly influenced by the composition of the soil microbiome close to roots. A strong correlation between the soil and root bacterial communities in A. thaliana has been reported by various authors [3,18]. Similarly, the structure of fungal communities is influenced more predominately by soil type than by host plant [98]. The type of soil influenced the rhizosphere bacterial microbiota composition in lettuce [99], oak [16], Arabidopsis [3], and maize [100].
Several studies have shown that environmental variability, such as soil pH, C: N ratio, soil carbon, water content and biogeography may influence the microbiota composition [39]. Lauber et al. [101] described that the impact of soil pH on total community composition was obvious even at a very high taxonomic level. Analyses revealed that pH has substantial correlation with the structure of these microbiome phyla in all soil types studied. The effects of soil pH on soil bacterial community composition has also been reported in other studies using various methods [102]. Zarraonaindia et al. [103] reported that the composition of soil and root microbiome of grapevine significantly influenced by soil pH and C:N ratio but leaf- and grape-associated microbiota were mostly influenced by soil carbon. Hartman, Richardson et al. [104] reported from their study that pH was the most important factor in predicting the alteration of soil bacterial communities, and they detected changes in phylum-level abundances across the pH levels. However, contrasting results have been reported by Fierer et al. [105], where they noted soil carbon was more important than soil pH for Bacteroidetes, Betaproteobacteria, and Acidobacteria. This difference of the findings was possibly linked to sample sizes and number of soil types investigated as well as to differences in methodologies. The exact mechanism(s) of the role of soil pH on microbial community composition and diversity is unknown. Two general explanations have been given [101]. Firstly, soil pH may indirectly alter bacterial community structure with its influences on soil characteristics. Secondly, soil pH may directly impose a physiological limit on soil bacteria, changing outcomes of competition or reducing survival of taxa intolerable to condition. In addition, we hypothesize that soil pH may alter plant microbiome by its influence on plant growth and physiology, which also may influence soil microbiome composition. Other soil properties, for example soil temperature and contaminants in the soil, also influence microbiome composition. Heat disturbance of soil results in a shift in rhizobacterial microbiome composition [106]. Heat disturbances due to natural wildfires can cause a decrease in microbial activity and significant alterations in microbial communities [107]. Increasing petroleum hydrocarbon contamination levels results in the alteration of willow microbiome structure. These alterations were less extreme in the rhizosphere and plant tissues, but they were prominent in the bulk soil. These could be because plants provide more controlled conditions and shield microbes against an enhanced contamination gradient [108].
8.2. Cultivation practices
Land use and cultivation practices are the most important causes of declines in biodiversity, leading to undesirable consequences for the environment [109]. Changes in the vegetation influence the diversity and structure of soil microbiomes. Agricultural activities do not necessarily have negative consequences in soil bacterial community diversity and structure, but they may have positive or neutral feedback (effect not perceived) [110]. For example, the intensity of land use (LUI) has been reported to influence the pattern of bacterial communities. Estendorfer et al. [111] found that under low LUI, there remains a strong interaction between plants and adjacent soil. In contrast, no influence of LUI on microbial diversity has been found in the rhizosphere, which indicates that plant species have much more influence on the rhizosphere community than soil properties do [112]. However, Suleiman et al. [113] reported that plants may have robust core microbiome compositions that are less prone to alteration due to variation of land use, soil type or edaphic factors. It was found that microbiome of in relatively untouched deciduous forest and long-term mowed grassland soils were comparable, although there were significant differences in soil properties and vegetation [114], Continuous cultivation causes changes in soil properties, which in turn might affect the soil microbiome communities. As per the reports of Allison and Martiny [115], there are three potential impacts caused by land disturbance such as the microbial structure might be affected, it might be changed but quickly return to the original composition (resilient), or might remain unchanged.
8.3. Climatic variables
Climate is an important driver of plant and soil microbiome composition. The role of climate in influencing plant microbiome community composition has to date been largely unheeded or found to be of little importance [116]. A recent study, however, across various ecosystems in Britain showed that rainfall and temperature gradients are two major climatic factors in shaping the bacterial community composition in plants [117]. Researchers de Vries et al. [118] reported that precipitation is an important driver of soil microbial community composition. The biomass of fungi and bacteria has been reported to increase with increasing mean annual precipitation (MAP), and the effect was more prominent for fungi, resulting in comparatively higher fungal abundance (increased F/B ratio) under higher precipitation levels. The above trends could be linked to higher soil organic matter contents in higher rainfall areas. In particular, in uplands where harsh climatic conditions prevail, a higher organic matter build-up is noticeable, which leads to fungi-dominated microbiome communities [119].
9. Conclusions and future perspective
The factors that influence microbiome assemblages and dynamics in plant and soil are now better understood, and research in this aspect is increasing. However, our knowledge on the underlying mechanism(s) of microbiome assemblages and how they influence the host plants is still lacking. Connecting the microbiome composition and diversity to their function is a great challenge for future research. For instance, to what extent could we use and manipulate the plant microbiome to boost sustainable agricultural production and environmental protection? We now know that that host genetic factor has a significant influence on microbiome diversity and structure, indicating that breeding and trait selection provide opportunities to select for desired microbiomes [45]. To have a more profound and broader knowledge on plant microbiome, there is a need to integrate novel molecular approaches (e.g., meta-omics), ecological models (e.g., food web theory, assembly of communities, or coexistence theory), and recent bioinformatics and statistical advances with a view to correlating community assemblages with ecological functions.
In the future, more emphasis should be placed on identifying the underlying mechanisms that drive microbiome community composition and assembly. We need to know the contributions of (1) the microbe-microbe interactions, (2) soil and other environmental variable and (3) various host traits in shaping community structure. Future research will direct toward solving some pertinent questions. For example, how stable are the drivers of microbiome community? To what extent do agricultural practices affect the microbiome of plant species? How predictable are these divers? With high throughput technologies, such as next-generation sequencing and metagenomics, we can begin to study endophyte microbiomes across hosts, environmental conditions, and at different time points and focus on the mechanisms of the plant-endophyte association.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Stephen J Wylie, PhD, Murdoch University, Australia for kindly checking the manuscript in part.
References
[1] G. Berg, D. Rybakova, M. Grube, M. Köberl, The plant microbiome explored: implications for experimental botany, J. Exp. Bot. 67 (2015) 995–1002. Google Scholar
[2] M. McFall-Ngai, M.G. Hadfield, T.C. Bosch, H.V. Carey, T. Domazet-Lošo, A.E. Douglas, et al., Animals in a bacterial world, a new imperative for the life sciences, Proc. Natl. Acad. Sci. U. S. A. 110 (2013) 3229–3236. Google Scholar
[3] D. Bulgarelli, M. Rott, K. Schlaeppi, E.V.L. van Themaat, N. Ahmadinejad, F. Assenza, et al., Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota, Nature 488 (2012) 91. Google Scholar
[4] R.L. Berendsen, C.M.J. Pieterse, P.A.H.M. Bakker, The rhizosphere microbiome and plant health, Trends Plant Sci. 17 (2012) 478–486. Google Scholar
[5] I. Zilber-Rosenberg, E. Rosenberg, Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution, FEMS Microbiol. Rev. 32 (2008) 723–735. Google Scholar
[6] R.T. Koide, B. Mosse, A history of research on arbuscular mycorrhiza, Mycorrhiza 14 (2004) 145–163. Google Scholar
[7] D.S. Heckman, D.M. Geiser, B.R. Eidell, R.L. Stauffer, N.L. Kardos, S.B. Hedges, Molecular evidence for the early colonization of land by fungi and plants, Science 293 (2001) 1129–1133. Google Scholar
[8] D. Bulgarelli, K. Schlaeppi, S. Spaepen, E.V.L. van Themaat, P. Schulze-Lefert, Structure and functions of the bacterial microbiota of plants, Annu. Rev. Plant Biol. 64 (2013) 807–838. Google Scholar
[9] T.R. Turner, E.K. James, P.S. Poole, The plant microbiome, Genome Biol. 14 (2013). Google Scholar
[10] G.D. Werner, W.K. Cornwell, J.I. Sprent, J. Kattge, E.T. Kiers, A single evolutionary innovation drives the deep evolution of symbiotic N 2-fixation in angiosperms, Nat. Commun. 5 (2014) 4087. Google Scholar
[11] M.G. Bakker, D.K. Manter, A.M. Sheflin, T.L. Weir, J.M. Vivanco, Harnessing the rhizosphere microbiome through plant breeding and agricultural management, Plant Soil 360 (2012) 1–13. Google Scholar
[12] P. Hinsinger, A.G. Bengough, D. Vetterlein, I.M. Young, Rhizosphere: biophysics, biogeochemistry and ecological relevance, Plant Soil 321 (2009) 117–152. Google Scholar
[13] M. Bonkowski, C. Villenave, B. Griffiths, Rhizosphere fauna: the functional and structural diversity of intimate interactions of soil fauna with plant roots, Plant Soil 321 (2009) 213–233. Google Scholar
[14] J. Gans, M. Wolinsky, J. Dunbar, Computational improvements reveal great bacterial diversity and high metal toxicity in soil, Science 309 (2005) 1387–1390. Google Scholar
[15] Ö İnceoğlu, W.A. Al-Soud, J.F. Salles, A.V. Semenov, J.D. van Elsas, Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing, PLoS One 6 (2011) e23321. Google Scholar
[16] S. Uroz, M. Buée, C. Murat, P. Frey‐Klett, F. Martin, Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil, Environ. Microbiol. Rep. 2 (2010) 281–288. Google Scholar
[17] D.S. Lundberg, S.L. Lebeis, S.H. Paredes, S. Yourstone, J. Gehring, S. Malfatti, et al., Defining the core Arabidopsis thaliana root microbiome, Nature 488 (2012) 86. Google Scholar
[18] K. Schlaeppi, N. Dombrowski, R.G. Oter, E.V.L. van Themaat, P. Schulze-Lefert, Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives, Proc. Natl. Acad. Sci. U. S. A. 111 (2014) 585–592. Google Scholar
[19] M.G. Van Der Heijden, F.M. Martin, M.A. Selosse, I.R. Sanders, Mycorrhizal ecology and evolution: the past, the present, and the future, New Phytol. 205 (2015) 1406–1423. Google Scholar
[20] M.K. Rich, E. Nouri, P.-E. Courty, D. Reinhardt, Diet of arbuscular mycorrhizal fungi: bread and butter? Trends Plant Sci. 22 (2017) 652–660. Google Scholar
[21] E.-H. Lee, J.-K. Eo, K.-H. Ka, A.-H. Eom, Diversity of arbuscular mycorrhizal fungi and their roles in ecosystems, Mycobiology 41 (2013) 121–125. Google Scholar
[22] S.E. Lindow, Role of Immigration and Other Processes in Determining Epiphytic Bacterial Populations. Aerial Plant Surface Microbiology, Springer, 1996, pp. 155–168. Google Scholar
[23] O.M. Finkel, A.Y. Burch, S.E. Lindow, A.F. Post, S. Belkin, Geographical location determines the population structure in phyllosphere microbial communities of a salt-excreting desert tree, Appl. Environ. Microbiol. 77 (2011) 7647–7655. Google Scholar
[24] J.A. Vorholt, Microbial life in the phyllosphere, Nat. Rev. Microbiol. 10 (2012) 828. Google Scholar
[25] S.E. Lindow, M.T. Brandl, Microbiology of the phyllosphere, Appl. Environ. Microbiol. 69 (2003) 1875–1883. Google Scholar
[26] N. Bodenhausen, M.W. Horton, J. Bergelson, Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana, PLoS One 8 (2013) e56329. Google Scholar
[27] D. Vokou, K. Vareli, E. Zarali, K. Karamanoli, H.-I.A. Constantinidou, N. Monokrousos, et al., Exploring biodiversity in the bacterial community of the Mediterranean phyllosphere and its relationship with airborne bacteria, Microb. Ecol. 64 (2012) 714–724. Google Scholar
[28] K.M.G. Dastogeer, H. Li, K. Sivasithamparam, M.G.K. Jones, S.J. Wylie, Host specificity of endophytic mycobiota of wild Nicotiana plants from arid regions of northern Australia, Microb. Ecol. (2017), https://doi.org/10.1007/s00248-017-1020-0. Google Scholar
[29] K.M. Dastogeer, H. Li, K. Sivasithamparam, M.G. Jones, S.J. Wylie, Fungal endophytes and a virus confer drought tolerance to Nicotiana benthamiana plants through modulating osmolytes, antioxidant enzymes and expression of host drought responsive genes, Environ. Exp. Bot. 149 (2018) 95–108. Google Scholar
[30] K.M.G. Dastogeer, H. Li, K. Sivasithamparam, M.G.K. Jones, X. Du, Y. Ren, et al., Metabolic responses of endophytic Nicotiana benthamiana plants experiencing water stress, Environ. Exp. Bot. 143 (2017) 59–71. Google Scholar
[31] R. Rodriguez, J. White Jr, A.E. Arnold, R.S. Redman, Fungal endophytes: diversity and functional roles, New Phytol. 182 (2009) 314–330. Google Scholar
[32] M.L. Bouffaud, M.A. Poirier, D. Muller, Y. Moenne-Loccoz, Root microbiome relates to plant host evolution in maize and other Poaceae, Environ. Microbiol. 16 (2014) 2804–2814. Google Scholar
[33] A. Samad, F. Trognitz, S. Compant, L. Antonielli, A. Sessitsch, Shared and host-specific microbiome diversity and functioning of grapevine and accompanying weed plants, Environ. Microbiol. 19 (2017) 1407–1424. Google Scholar
[34] D.K. Manter, J.A. Delgado, D.G. Holm, R.A. Stong, Pyrosequencing reveals a highly diverse and cultivar-specific bacterial endophyte community in potato roots, Microb. Ecol. 60 (2010) 157–166. Google Scholar
[35] S.W. Kembel, T.K. O’Connor, H.K. Arnold, S.P. Hubbell, S.J. Wright, J.L. Green, Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest, Proc. Natl. Acad. Sci. U. S. A. 111 (2014) 13715–13720. Google Scholar
[36] C.R. Fitzpatrick, J. Copeland, P.W. Wang, D.S. Guttman, P.M. Kotanen, M.T. Johnson, Assembly and ecological function of the root microbiome across angiosperm plant species, Proc. Natl. Acad. Sci. U. S. A. 115 (2018) E1157–E1165. Google Scholar
[37] X. Xiao, W.M. Chen, L. Zong, J. Yang, S. Jiao, Y.B. Lin, et al., Two cultivated legume plants reveal the enrichment process of the microbiome in the rhizocompartments, Mol. Ecol. 26 (2017) 1641–1651. Google Scholar
[38] D. Bulgarelli, R. Garrido-Oter, P.C. Münch, A. Weiman, J. Dröge, Y. Pan, et al., Structure and function of the bacterial root microbiota in wild and domesticated barley, Cell Host Microbe 17 (2015) 392–403. Google Scholar
[39] J.A. Peiffer, A. Spor, O. Koren, Z. Jin, S.G. Tringe, J.L. Dangl, et al., Diversity and heritability of the maize rhizosphere microbiome under field conditions, Proc. Natl. Acad. Sci. U. S. A. (2013) 201302837. Google Scholar
[40] R.A. Ortega, A. Mahnert, C. Berg, H. Muller, G. Berg, The plant is crucial: specific composition and function of the phyllosphere microbiome of indoor ornamentals, FEMS Microbiol. Ecol. 92 (2016). Google Scholar
[41] J.M. Marques, T.F. da Silva, R.E. Vollu, A.F. Blank, G.-C. Ding, L. Seldin, et al., Plant age and genotype affect the bacterial community composition in the tuber rhizosphere of field-grown sweet potato plants, FEMS Microbiol. Ecol. 88 (2014)
424–435. Google Scholar
[42] S. Hacquard, C.W. Schadt, Towards a holistic understanding of the beneficial interactions across the Populus microbiome, New Phytol. 205 (2015) 1424–1430. Google Scholar
[43] M.W. Horton, N. Bodenhausen, K. Beilsmith, D. Meng, B.D. Muegge, S. Subramanian, et al., Genome-wide association study of Arabidopsis thaliana leaf microbial community, Nat. Commun. 5 (2014) 5320. Google Scholar
[44] M.R. Wagner, D.S. Lundberg, G. Tijana, S.G. Tringe, J.L. Dangl, T. Mitchell-Olds, Host genotype and age shape the leaf and root microbiomes of a wild perennial plant, Nat. Commun. 7 (2016) 12151. Google Scholar
[45] M.T. Agler, J. Ruhe, S. Kroll, C. Morhenn, S.T. Kim, D. Weigel, et al., Microbial hub taxa link host and abiotic factors to plant microbiome variation, PLoS Biol. 14 (2016) e1002352. Google Scholar
[46] F. Cai, G. Pang, Y.Z. Miao, R.X. Li, R.X. Li, Q.R. Shen, et al., The nutrient preference of plants influences their rhizosphere microbiome, Appl. Soil Ecol. 110 (2017) 146–150. Google Scholar
[47] J. Edwards, C. Johnson, C. Santos-Medellín, E. Lurie, N.K. Podishetty, S. Bhatnagar, et al., Structure, variation, and assembly of the root-associated microbiomes of rice, Proc. Natl. Acad. Sci. U. S. A. 112 (2015) E911–E920. Google Scholar
[48] D. Coleman-Derr, D. Desgarennes, C. Fonseca-Garcia, S. Gross, S. Clingenpeel, T. Woyke, et al., Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species, New Phytol. 209 (2016) 798–811. Google Scholar
[49] J.W. Taylor, E. Turner, J.P. Townsend, J.R. Dettman, D. Jacobson, Eukaryotic microbes, species recognition and the geographic limits of species: examples from the kingdom Fungi, Philos. Trans. R. Soc. Lond. B: Biol. Sci. 361 (2006) 1947–1963. Google Scholar
[50] P. Bednarek, Chemical warfare or modulators of defence responses–the function of secondary metabolites in plant immunity, Curr. Opin. Plant Biol. 15 (2012) 407–414. Google Scholar
[51] A. Sugiyama, Y. Ueda, T. Zushi, H. Takase, K. Yazaki, Changes in the bacterial community of soybean rhizospheres during growth in the field, PLoS One 9 (2014) e100709. Google Scholar
[52] J.M. Chaparro, D.V. Badri, J.M. Vivanco, Rhizosphere microbiome assemblage is affected by plant development, ISME J. 8 (2014) 790–803. Google Scholar
[53] A. Lerner, Y. Herschkovitz, E. Baudoin, S. Nazaret, Y. Moenne-Loccoz, Y. Okon, et al., Effect of Azospirillum brasilense inoculation on rhizobacterial communities analyzed by denaturing gradient gel electrophoresis and automated ribosomal
intergenic spacer analysis, Soil Biol. Biochem. 38 (2006) 1212–1218. Google Scholar
[54] Y. Xu, G. Wang, J. Jin, J. Liu, Q. Zhang, X. Liu, Bacterial communities in soybean rhizosphere in response to soil type, soybean genotype, and their growth stage, Soil Biol. Biochem. 41 (2009) 919–925. Google Scholar
[55] L. Augusto, J. Ranger, D. Binkley, A. Rothe, Impact of several common tree species of European temperate forests on soil fertility, Ann. For. Sci. 59 (2002) 233–253. Google Scholar
[56] J. Monaghan, C. Zipfel, Plant pattern recognition receptor complexes at the plasma membrane, Curr. Opin. Plant Biol. 15 (2012) 349–357. Google Scholar
[57] M. Erb, C. Lenk, J. Degenhardt, T.C. Turlings, The underestimated role of roots in defense against leaf attackers, Trends Plant Sci. 14 (2009) 653–659. Google Scholar
[58] B. Lee, S. Lee, C.-M. Ryu, Foliar aphid feeding recruits rhizosphere bacteria and primes plant immunity against pathogenic and non-pathogenic bacteria in pepper, Ann. Bot. 110 (2012) 281–290. Google Scholar
[59] Y. Chen, F. Yan, Y. Chai, H. Liu, R. Kolter, R. Losick, et al., Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation, Environ. Microbiol. 15 (2013)
848–864. Google Scholar
[60] V. Lakshmannan, S. Kitto, J. Caplan, Y.-H. Hsueh, D. Kearns, Y.-S. Wu, et al., Microbe-Associated Molecular Patterns (MAMPs)-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis, Plant Physiol. (2012) pp.
112.200386. Google Scholar
[61] R.F. Doornbos, B.P. Geraats, E.E. Kuramae, L. Van Loon, P.A. Bakker, Effects of jasmonic acid, ethylene, and salicylic acid signaling on the rhizosphere bacterial community of Arabidopsis thaliana, Mol. Plant-Microbe Interact. 24 (2011)
395–407. Google Scholar
[62] R. Landgraf, S. Schaarschmidt, B. Hause, Repeated leaf wounding alters the colonization of Medicago truncatula roots by beneficial and pathogenic microorganisms, Plant Cell Environ. 35 (2012) 1344–1357. Google Scholar
[63] S. Lebeis, A.C. McHardy, J.L. Dangl, R. Knight, R. Ley, P. Schulze-Lefert, Microbiota and host nutrition across plant and animal kingdoms, Cell Host Microbe 17 (2015) 603616. Google Scholar
[64] J.M. Kniskern, M.B. Traw, J. Bergelson, Salicylic acid and jasmonic acid signaling defense pathways reduce natural bacterial diversity on Arabidopsis thaliana, Mol. Plant-Microbe Interact. 20 (2007) 1512–1522. Google Scholar
[65] S. Hassan, U. Mathesius, The role of flavonoids in root–rhizosphere signalling: opportunities and challenges for improving plant–microbe interactions, J. Exp. Bot. 63 (2012) 3429–3444. Google Scholar
[66] K. Akiyama, H. Hayashi, Strigolactones: chemical signals for fungal symbionts and parasitic weeds in plant roots, Ann. Bot. 97 (2006) 925–931. Google Scholar
[67] A.G. Darvill, P. Albersheim, Phytoalexins and their elicitors-a defense against microbial infection in plants, Annu. Rev. Plant Physiol. 35 (1984) 243–275. Google Scholar
[68] P. Bednarek, A. Osbourn, Plant-microbe interactions: chemical diversity in plant defense, Science 324 (2009) 746–748. Google Scholar
[69] M. Bressan, M.-A. Roncato, F. Bellvert, G. Comte, F. el Zahar Haichar, W. Achouak, et al., Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots, ISME J. 3 (2009) 1243. Google Scholar
[70] J. Maizel, H. Burkhardt, H. Mitchell, Avenacin, an antimicrobial substance isolated from Avena sativa. I. Isolation and antimicrobial activity, Biochemistry 3 (1964) 424–426. Google Scholar
[71] J.P. Carter, J. Spink, P.F. Cannon, M.J. Daniels, A.E. Osbourn, Isolation, characterization, and avenacin sensitivity of a diverse collection of cereal-root-colonizing fungi, Appl. Environ. Microbiol. 65 (1999) 3364–3372. Google Scholar
[72] K. Papadopoulou, R. Melton, M. Leggett, M. Daniels, A. Osbourn, Compromised disease resistance in saponin-deficient plants, Proc. Natl. Acad. Sci. U. S. A. 96 (1999) 12923–12928. Google Scholar
[73] T.R. Turner, K. Ramakrishnan, J. Walshaw, D. Heavens, M. Alston, D. Swarbreck, et al., Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants, ISME J. 7 (2013) 2248. Google Scholar
[74] G. Harmsen, G. Jager, J. Doeksen, J. van der Drift (Eds.), Determination of the Quantity of Carbon and Nitrogen in the Rhizosphere of Young Plants, 1963, pp. 245–251 North Holland Amsterdam. Google Scholar
[75] R.O. Schlechter, M. Miebach, M.N. Remus-Emsermann, Driving factors of epiphytic bacterial communities: a review, J. Adv. Res. (2019). Google Scholar
[76] S. Banerjee, K. Schlaeppi, M.G. Heijden, Keystone taxa as drivers of microbiome structure and functioning, Nat. Rev. Microbiol. (2018) 1. Google Scholar
[77] S.A. Shetty, F. Hugenholtz, L. Lahti, H. Smidt, W.M. de Vos, Intestinal microbiome landscaping: insight in community assemblage and implications for microbial modulation strategies, FEMS Microbiol. Rev. 41 (2017) 182–199. Google Scholar
[78] J.M. Raaijmakers, D.M. Weller, Natural plant protection by 2, 4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils, Mol. Plant-Microbe Interact. 11 (1998) 144–152. Google Scholar
[79] N. Mathivanan, V. Prabavathy, V. Vijayanandraj, The effect of fungal secondary metabolites on bacterial and fungal pathogens, Secondary Metabolites in Soil Ecology, Springer, 2008, pp. 129–140. Google Scholar
[80] X.S. Ramírez-Gómez, S.N. Jiménez-García, V.B. Campos, M.L.G. Campos, Plant metabolites in plant defense against pathogens, Plant Pathology and Management of Plant Diseases, IntechOpen, 2019. Google Scholar
[81] C.M. Pieterse, C. Zamioudis, R.L. Berendsen, D.M. Weller, S.C. Van Wees, P.A. Bakker, Induced systemic resistance by beneficial microbes, Annu. Rev. Phytopathol. 52 (2014) 347–375. Google Scholar
[82] R.T. Voegele, M. Hahn, G. Lohaus, T. Link, I. Heiser, K. Mendgen, Possible roles for mannitol and mannitol dehydrogenase in the biotrophic plant pathogen Uromyces fabae, Plant Physiol. 137 (2005) 190–198. Google Scholar
[83] C. Poza-Carrion, T. Suslow, S. Lindow, Resident bacteria on leaves enhance survival of immigrant cells of Salmonella enterica, Phytopathology 103 (2013) 341–351. Google Scholar
[84] E. Roberts, S. Lindow, Loline alkaloid production by fungal endophytes of Fescue species select for particular epiphytic bacterial microflora, ISME J. 8 (2014) 359. Google Scholar
[85] N.R. Horner, L.J. Grenville-Briggs, P. Van West, The oomycete Pythium oligandrum expresses putative effectors during mycoparasitism of Phytophthora infestans and is amenable to transformation, Fungal Biol. 116 (2012) 24–41. Google Scholar
[86] M. McMullan, A. Gardiner, K. Bailey, E. Kemen, B.J. Ward, V. Cevik, et al., Evidence for suppression of immunity as a driver for genomic introgressions and host range expansion in races of Albugo candida, a generalist parasite, Elife 4 (2015) e04550. Google Scholar
[87] K. Buddrus-Schiemann, M. Schmid, K. Schreiner, G. Welzl, A. Hartmann, Root colonization by Pseudomonas sp. DSMZ 13134 and impact on the indigenous rhizosphere bacterial community of barley, Microb. Ecol. 60 (2010) 381–393. Google Scholar
[88] S.P. Chowdhury, K. Dietel, M. Rändler, M. Schmid, H. Junge, R. Borriss, et al., Effects of Bacillus amyloliquefaciens FZB42 on lettuce growth and health under pathogen pressure and its impact on the rhizosphere bacterial community, PLoS One 8 (2013) e68818. Google Scholar
[89] O.S. Correa, M.S. Montecchia, M.F. Berti, M.C.F. Ferrari, N.L. Pucheu, N.L. Kerber, et al., Bacillus amyloliquefaciens BNM122, a potential microbial biocontrol agent applied on soybean seeds, causes a minor impact on rhizosphere and soil microbial communities, Appl. Soil Ecol. 41 (2009) 185–194. Google Scholar
[90] A. Erlacher, M. Cardinale, R. Grosch, M. Grube, G. Berg, The impact of the pathogen Rhizoctonia solani and its beneficial counterpart Bacillus amyloliquefaciens on the indigenous lettuce microbiome, Front. Microbiol. 5 (2014) 175. Google Scholar
[91] M. Kröber, D. Wibberg, R. Grosch, F. Eikmeyer, B. Verwaaijen, S.P. Chowdhury, et al., Effect of the strain Bacillus amyloliquefaciens FZB42 on the microbial community in the rhizosphere of lettuce under field conditions analyzed by whole metagenome sequencing, Front. Microbiol. 5 (2014) 252. Google Scholar
[92] E. Fonseca, R. Peixoto, A. Rosado, F. Balieiro, J. Tiedje, C. Rachid, The microbiome of Eucalyptus roots under different management conditions and its potential for biological nitrogen fixation, Microb. Ecol. (2017). Google Scholar
[93] F. Valladares, E. Gianoli, J.M. Gómez, Ecological limits to plant phenotypic plasticity, New Phytol. 176 (2007) 749–763. Google Scholar
[94] F. Tardieu, Plant response to environmental conditions: assessing potential production, water demand, and negative effects of water deficit, Front. Physiol. 4 (2013) 17. Google Scholar
[95] T.R. Williams, M.L. Marco, Phyllosphere microbiota composition and microbial community transplantation on lettuce plants grown indoors, Mbio 5 (2014) e01564–14. Google Scholar
[96] B.K. Whitaker, H.L. Reynolds, K. Clay, Foliar fungal endophyte communities are structured by environment but not host ecotype in Panicum virgatum (switchgrass), Ecology 99 (2018) 2703–2711. Google Scholar
[97] N. Dombrowski, K. Schlaeppi, M.T. Agler, S. Hacquard, E. Kemen, R. Garrido-Oter, et al., Root microbiota dynamics of perennial Arabis alpina are dependent on soil residence time but independent of flowering time, ISME J. 11 (2017) 43–55. Google Scholar
[98] G. Bonito, H. Reynolds, M.S. Robeson, J. Nelson, B.P. Hodkinson, G. Tuskan, et al., Plant host and soil origin influence fungal and bacterial assemblages in the roots of woody plants, Mol. Ecol. 23 (2014) 3356–3370. Google Scholar
[99] S. Schreiter, G.-C. Ding, H. Heuer, G. Neumann, M. Sandmann, R. Grosch, et al., Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce, Front. Microbiol. 5 (2014) 144. Google Scholar
[100] A.B. Dohrmann, M. Küting, S. Jünemann, S. Jaenicke, A. Schlüter, C.C. Tebbe, Importance of rare taxa for bacterial diversity in the rhizosphere of Bt-and conventional maize varieties, ISME J. 7 (2013) 37. Google Scholar
[101] C.L. Lauber, M. Hamady, R. Knight, N. Fierer, Soil pH as a predictor of soil bacterial community structure at the continental scale: a pyrosequencing-based assessment, Appl. Environ. Microbiol. (2009). Google Scholar
[102] R. Upchurch, C.-Y. Chiu, K. Everett, G. Dyszynski, D.C. Coleman, W.B. Whitman, Differences in the composition and diversity of bacterial communities from agricultural and forest soils, Soil Biol. Biochem. 40 (2008) 1294–1305. Google Scholar
[103] I. Zarraonaindia, S.M. Owens, P. Weisenhorn, K. West, J. Hampton-Marcell, S. Lax, et al., The soil microbiome influences grapevine-associated microbiota, MBio 6 (2015) e02527–14. Google Scholar
[104] W.H. Hartman, C.J. Richardson, R. Vilgalys, G.L. Bruland, Environmental and anthropogenic controls over bacterial communities in wetland soils, Proc. Natl. Acad. Sci. U. S. A. (2008) pnas. 0808254105. Google Scholar
[105] N. Fierer, M.A. Bradford, R.B. Jackson, Toward an ecological classification of soil bacteria, Ecology 88 (2007) 1354–1364. Google Scholar
[106] M. van der Voort, M. Kempenaar, M. van Driel, J.M. Raaijmakers, R. Mendes, Impact of soil heat on reassembly of bacterial communities in the rhizosphere microbiome and plant disease suppression, Ecol. Lett. 19 (2016) 375–382. Google Scholar
[107] W.M. Jolly, M.A. Cochrane, P.H. Freeborn, Z.A. Holden, T.J. Brown, G.J. Williamson, et al., Climate-induced variations in global wildfire danger from 1979 to 2013, Nat. Commun. 6 (2015) 7537. Google Scholar
[108] S. Tardif, É Yergeau, J. Tremblay, P. Legendre, L.G. Whyte, C.W. Greer, The willow microbiome is influenced by soil petroleum-hydrocarbon concentration with plant compartment-specific effects, Front. Microbiol. 7 (2016) 1363. Google Scholar
[109] A.A. Navarrete, F.S. Cannavan, R.G. Taketani, S.M. Tsai, A molecular survey of the diversity of microbial communities in different Amazonian agricultural model systems, Diversity 2 (2010) 787–809. Google Scholar
[110] B.K. Singh, P. Millard, A.S. Whiteley, J.C. Murrell, Unravelling
rhizosphere–microbial interactions: opportunities and limitations, Trends Microbiol. 12 (2004) 386–393. Google Scholar
[111] J. Estendorfer, B. Stempfhuber, P. Haury, G. Vestergaard, M.C. Rillig, J. Joshi, et al., The influence of land use intensity on the plant-associated microbiome of Dactylis glomerata L, Front. Plant Sci. 8 (2017) 930. Google Scholar
[112] N.R. Gottel, H.F. Castro, M. Kerley, Z. Yang, D.A. Pelletier, M. Podar, et al., Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types, Appl. Environ. Microbiol. 77 (2011) 5934–5944. Google Scholar
[113] A.K.A. Suleiman, V.S. Pylro, L.F.W. Roesch, Replacement of native vegetation alters the soil microbial structure in the Pampa biome, Sci. Agric. 74 (2017) 77–84. Google Scholar
[114] K. Jangid, M.A. Williams, A.J. Franzluebbers, T.M. Schmidt, D.C. Coleman, W.B. Whitman, Land-use history has a stronger impact on soil microbial community composition than aboveground vegetation and soil properties, Soil Biol. Biochem. 43 (2011) 2184–2193. Google Scholar
[115] S.D. Allison, J.B. Martiny, Resistance, resilience, and redundancy in microbial communities, Proc. Natl. Acad. Sci. U. S. A. 105 (2008) 11512–11519. Google Scholar
[116] N. Fierer, R.B. Jackson, The diversity and biogeography of soil bacterial communities, Proc. Natl. Acad. Sci. U. S. A. 103 (2006) 626–631. Google Scholar
[117] R.I. Griffiths, B.C. Thomson, P. James, T. Bell, M. Bailey, A.S. Whiteley, The bacterial biogeography of British soils, Environ. Microbiol. 13 (2011) 1642–1654. Google Scholar
[118] F.T. de Vries, P. Manning, J.R. Tallowin, S.R. Mortimer, E.S. Pilgrim, K.A. Harrison, et al., Abiotic drivers and plant traits explain landscape‐scale patterns in soil microbial communities, Ecol. Lett. 15 (2012) 1230–1239. Google Scholar
[119] R.D. Bardgett, A.C. Jones, D.L. Jones, S.J. Kemmitt, R. Cook, P.J. Hobbs, Soil microbial community patterns related to the history and intensity of grazing in sub-montane ecosystems, Soil Biol. Biochem. 33 (2001) 1653–1664.
K.M.G. Dastogeer, et al. Current Plant Biology 23 (2020) 100161 Google Scholar
Hotspot Quiz
Check your Understanding
- What are the various types of plant-associated microbiomes and how does each affect plant health?
- How are the compositions of plant microbiomes influenced by the environment?
- What aspects of the host plant can shape its microbiome?
- In what ways can the plant microbiome be governed by microbe-microbe interactions?
Media Attributions
- Video 1 – Disease-induced changes in plant microbiome assembly and functional adaptation by Research Square licensed under Creative Commons Attribution license (reuse allowed)
- Video 2 – [microbiome] The Plant Microbiome: The plant microbiome in facts (3.1) by iMooX at licenced under Creative Commons Attribution-ShareAlike 4.0 International.
- Video 3 – [microbiome] The Plant Microbiome: The apple microbiome (3.2) by iMooX at licenced under Creative Commons Attribution-ShareAlike 4.0 International.
- Video 4 – [microbiome] The Plant Microbiome: Managing the plant microbiome (3.3) by iMooX at licenced under Creative Commons Attribution-ShareAlike 4.0 International.
References
-
Dastogeer, K. M. G., Tumpa, F. H., Sultana, A., Akter, M. A., & Chakraborty, A. (2020). Plant microbiome–an account of the factors that shape community composition and diversity. Current Plant Biology, 23, 100161. https://doi.org/10.1016/j.cpb.2020.100161

