1 An Introduction to Microbiomes
An Introduction to Microbiomes
Microorganisms represent the fundamentals of life and interact with almost every facet of it. Yet, for the majority of their existence they have been largely ignored, or rather unseen by humans. The unraveling of the complex interactions between all life forms is an endless duty and seems to generate more questions than answers. However, the cracks in knowledge produced by peering into the unknown offers insight into our life and the world around us. These minute creatures have shaped Earth’s evolution since their dawn almost three-and-a-half billion years ago, and continuously affect our environment and health. The importance of the planet’s collection of microbes is realized more and more each day, and our unceasing investigation of them will surely unlock secrets of life we could never imagine.
The study of microbiomes is fairly novel in the context of understanding and applying their communal existence to ourselves and surroundings. Though scientists have recognized symbiotic relationships and traditionally focused on individual microbes and their interactions with human health, environmental impact, industrial applications, etc., their respective communities and influence as a whole in these areas have only just begun to be elucidated. That is in no small part due to the daunting task of cataloging the immense and complex interplay between the multitude of different microorganisms in a given environment, though rapid advances in technology have begun to ease analysis.
What is a microbiome?
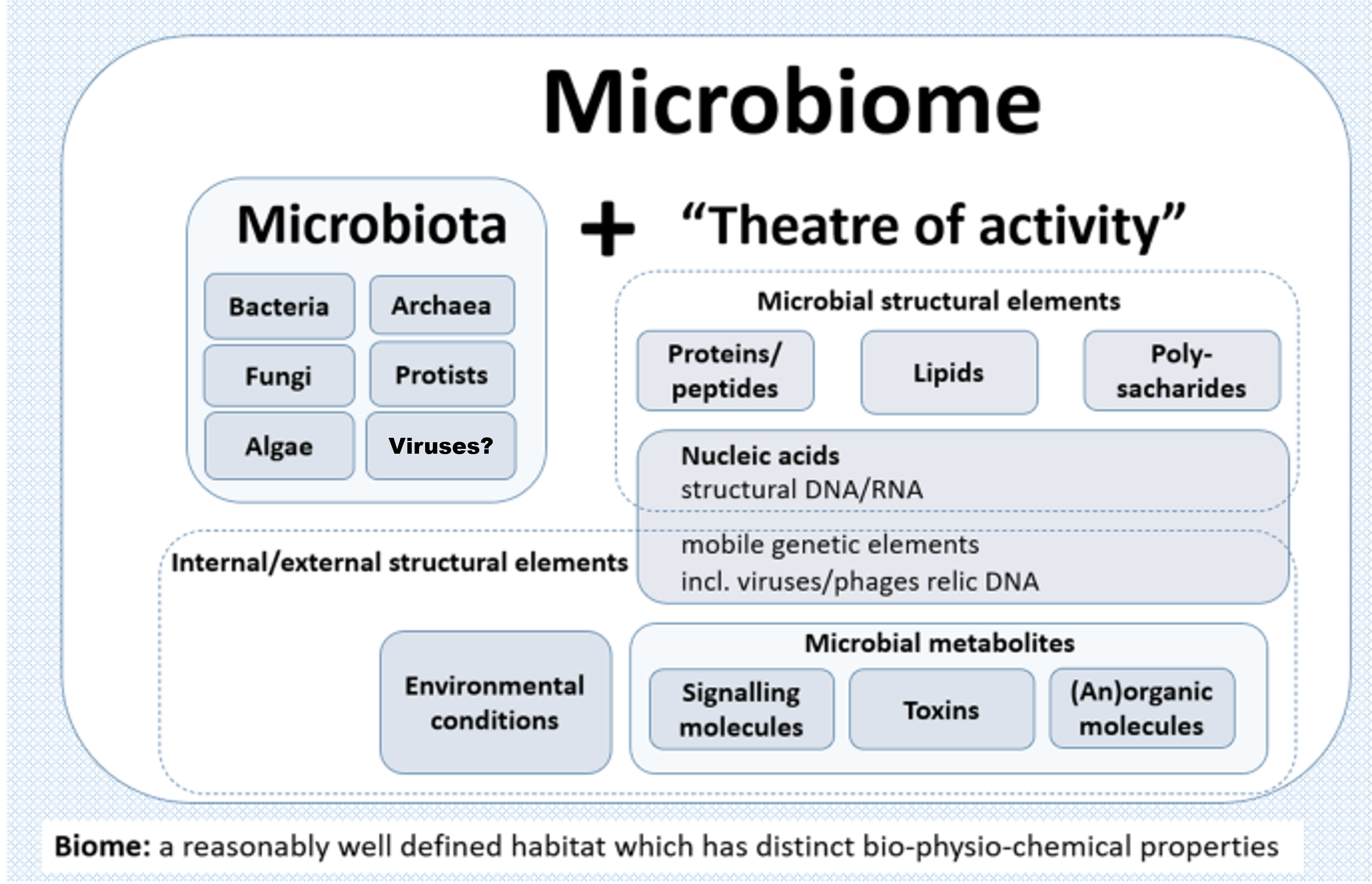
A microbiome can be best described as a collective polymicrobial community, or ‘microbiota’, and its associated activity with genetic and physio-chemical constituents in a defined spaciotemporal habitat (Figure 1). These members of the microbiota include bacteria, archaea, algae, protozoa, fungi, and viruses (though the latter is somewhat debated since viruses and their derivatives aren’t technically living). Within this symbiotic context with a particular eukaryotic host, the entire entity is termed a ‘holobiont’ and the aggregate of genetic material termed the ‘hologenome’. Interactions between these partners may have long occurred, shaping the evolution of each, whereas others may be novel or transient, sometimes resulting in prompt change and infectious diseases. The change in the normal microbiota, or dysbiosis, can result in a variety of different diseases. As so, their study has been especially important in the fields of life sciences, human health, and medicine.
Quick Quiz
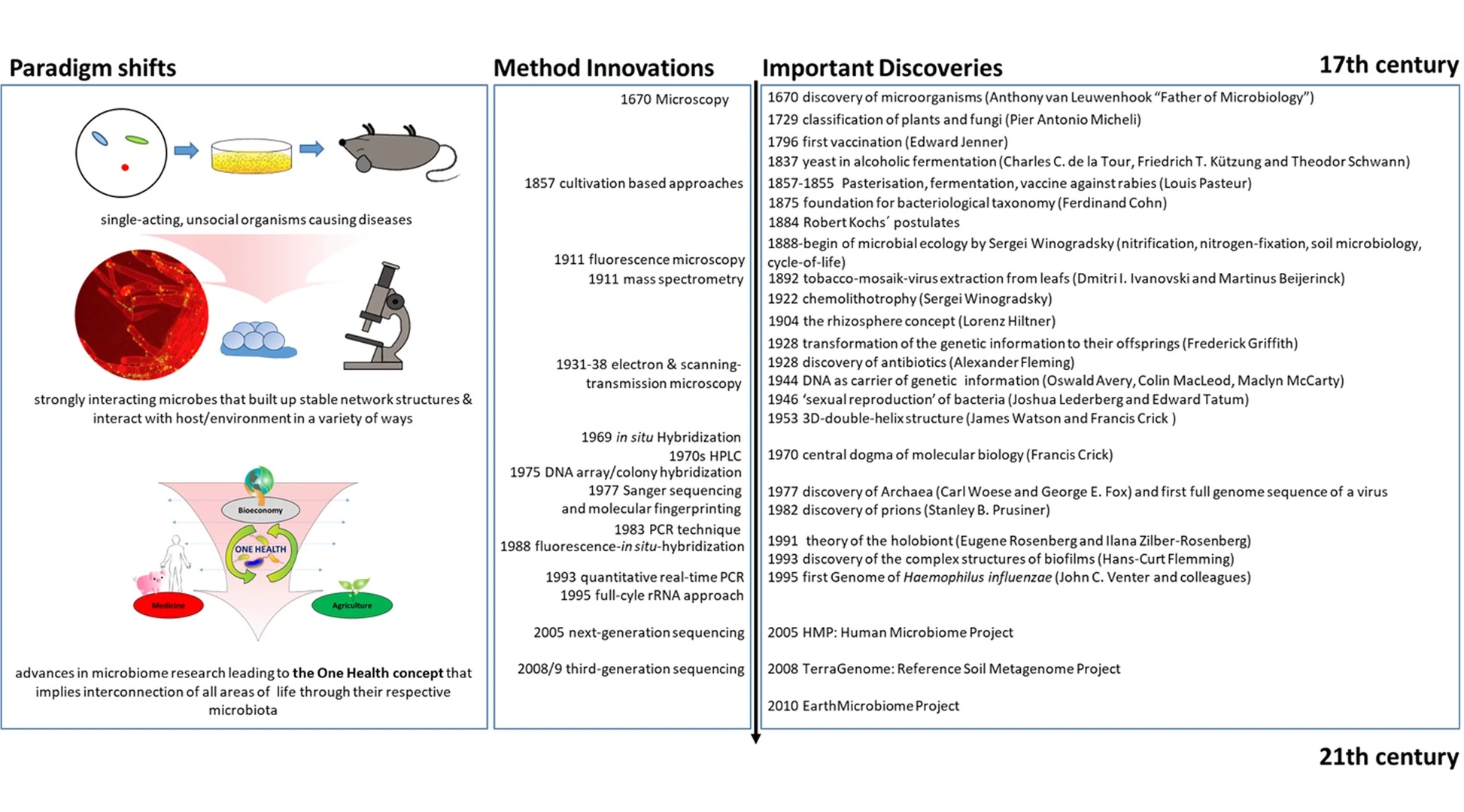
Our fascination with microorganisms began before we even fully understood them or were even able to see them. Their implications concerning human health were primarily explored during the ‘golden age of microbiology’ with the work of Louis Pasteur and Robert Koch. Their experiments and discoveries shed light on not only the ubiquitous nature of microbes, but their importance in our everyday lives. Other significant milestones and historical microbiological development can be viewed in Figure 2. Human health and infectious diseases were central to the field of microbiology, though food microbiology, industrial applications, and microbial ecology became increasingly explored. Over the last couple centuries, a microbial catalog of knowledge has slowly grown, but much of these findings were limited to those organisms that could be cultured and measured. The advent of sequencing and ‘multi-omics’ technologies has since allowed researchers to document microorganisms that were previously missed or ignored with traditional techniques, and with further advances, larger microbial communities and symbioses can be better understood. The ‘microbiome’ was first defined in the late 1980s when a group of microbial ecologists were studying the rhizosphere, which provided context to better describe these polymicrobial communities (Whipps et al., 1988). Many other similar definitions have been published since then with varying specifics on genetic expression, symbioses, and ecological interactions (Lederberg & McCray, 2001, Marchesi & Ravel, 2015, Berg et al., 2020). The ‘holobiont’ concept stems from Adolf Meyer-Abich’s ‘theory of holobiosis’ proposed in 1943 and was independently conceived and popularized in the early 1990s by Lynn Margulis, though it only described the host and a single symbiont (Margulis, 1991, Baedke et al., 2020). Since then has been expanded to include the entire microbiota in multiple symbiotic contexts (Simon et al., 2019). In recent decades there has been a steady increase in microbiome publications as the subject has grown in popularity. Along with that, there has been more analytical breakdown as certain microbiomes are being described with emphasis on specific members, such as the ‘bacteriome’, ‘archaeome’, ‘mycobiome’, ‘protistome’, and ‘virome’, and these terms are best used to refer to the distinct contribution of those particular microbes within the entire microbiome context. In general, though, most microbiomes are delineated by their specific host or type of environment, with the human microbiome being the most popular example.
The Human Microbiome
It was evident that the human microbiome and its involvement in a micro and macro scale needed to be characterized. The Human Microbiome Project (HMP) set out in 2007 with this as one of its primary goals (Turnbaugh et al., 2007). The program also set out with initiatives to develop a set of microbial genome sequences, explain the relationship between disease and microbiome changes and evaluate the data with multi-omics approaches, develop new tools and technology for computational analysis, establish a data analysis and coordinating center and research repositories, as well as address ethical, social, and legal implications of HMP research (Human Microbiome Project). The second phase of the HMP launched in 2014, called the Integrative Human Microbiome Project (iHMP), having the main mission to completely characterize the human microbiota with a key focus on human health and disease using three projects: pregnancy and preterm birth, onset of inflammatory bowel disease (IBD), and onset of type 2 diabetes (NIH Human Microbiome Project, The Integrative HMP (iHMP) Research Network Consortium, 2019). Aside from these, the human microbiome and disruption of the microbiota has been linked to several other important conditions and diseases including multiple sclerosis, diabetes (types 1 and 2), allergies, asthma, autism, and cancer (Backhed et al., 2012, Hsiao et al., 2013, Petersen and Round, 2014, Trompette et al., 2014, Garrett, 2015, Lloyd-Price et al., 2016).
It makes sense that the human microbiome can have such an impact on human health and behavior if you consider that we are essentially a collection of organisms forming a living entity. In a way, our symbionts may even actually define more of who we are than just our own unique biological makeup. For instance, the ratio of microbial cells associated with a human body could equal, if not exceed (traditional estimates were tenfold), the number of human cells (Sender et al., 2016). Even more interesting is viewing our genetic makeup; the human genome contains about 20,000 genes, but its hologenome contains > 33 million genes brought by its microbiota (Huttenhower et al., 2012, Lloyd-Price et al., 2016, Simon et al., 2019). Furthermore, the composition and rate of change of each person’s microbiota is distinctive from one individual to another since it is influenced by variables like age, lifestyle, diet, antibiotics, occupation, environment, etc. (Gilbert et al., 2018). The genetic wealth and member diversity contributed from the microbiota has roles in adaptation, survival, development, growth, and reproduction of the holobiont and can affect fitness in the short term as well as have long lasting effects concerning the evolution of both partners (Rosenberg, and Zilber-Rosenberg, 2011).
Co-evolution of the host-microbiota symbiosis can be considered even more unique when viewing the microbial consortium at different locations or organs in the host, as their makeup is governed by and reflects specific physiological processes in those areas. For example, the bacteria found in the human gut microbiota are primarily from the phyla Bacteroides and Firmicutes, whereas Actinobacteria and Proteobacteria command the skin microbiome, though there is some overlap and it is important to note that there are differences depending on exact location (e.g. dry vs. moist areas of the skin) (Grice and Segre, 2011, Jandhyala et al. 2015). Though there are differences between various microbiota within a holobiont, they can still influence each other to some degree. In the case of the gut and skin microbiotas in humans, deemed the ‘gut-skin axis’, there are indications that both the health of the gastrointestinal (GI) tract and skin, as well as their response to stressors, are correlated (Levcovich et al., 2013, O’Neill et al., 2016, Salem et al. 2018). Even more interesting is the effects certain microbiota can have on germ-free organs like the brain. Studies on the ‘gut-brain axis’ show that the microbiota in the GI tract, and in some cases disruption of it, are associated with many mental illnesses and neurodegenerative disorders including depression, anxiety, autism, schizophrenia, Parkinson’s disease, and Alzheimer’s disease (Clapp et al. 2017, Foster et al. 2017, Cryan et al. 2019). A variety of different ‘axes’ which demonstrate interplay between microbiota, organs, and locations have been identified in the human body and much of what is known about their connections is novel and early in its research.
Environmental Microbiomes
Not only can our microbiome regulate who we are, but those communities in the surrounding environment can affect us, our microbiome, and others. Environmental microbiomes can directly or indirectly affect our health through ecological interactions. For example, soil microbiomes in the rhizosphere of plant roots and plant microbiomes of economically important crops have implications in agriculture, human health, and ecology (Saleem et al., 2019, Hirt, 2020). Plant growth, health, soil nutrient cycling and availability, and defense against potential pathogens are dictated by their own symbionts as well as their microbial neighbors in the ground, which include a variety of bacteria, protists, viruses, and network of fungi known as mycorrhizae (Busby et al., 2017, Hannula et al., 2017, Pratama and van Elsas, 2018, Zhong et al., 2019,). By better understanding the functional interactions of all the players in these environmental microbiomes, it may be possible to address problems like agricultural soil fertility, plant disease, and pollution. Thus, harnessing better microbiomes either by specifically engineering a microbial consortia for a targeted area/specimen or transplanting a natural community could improve sustainable agricultural practices which are desperately needed to feed the expanding population of humans while protecting the environment (Elhady et al., 2018, Arif et al., 2020, Hernandez-Alvarez et al., 2022).
Studying animal-microbiome systems can also give us information about our environment. For instance, how anthropogenic-induced changes have impacted things like climate-change and approaches to maintain homeostasis with various environmental factors. A promising reservoir of research is the microbiota of marine animals since most of the planet is covered in water. Corals, sponges, various fish, and marine mammals have all been investigated to document the response of their microbial symbionts to changing environmental factors (Apprill, 2017). Other animal microbiome models are being used in an analogous means to understand how the microbiota could potentially influence human health and fitness. The microbiomes of classic biomedical models such as the fruit fly, zebrafish, and nematode worm are being investigated since these organisms are well known and readily available to work with in many labs (Douglas, 2019). Though, it is important to consider a variety of animal host-microbiome models as each could contribute unique insights to beneficial microbial interactions within the human holobiont. For example, the gut microbiome of honeybees, the skin microbiome of freshwater polyps, and the individual interaction of Vibro fischeri and the Hawaiian bob-tailed squid have provided valuable information to understanding host-microbiome relationships (Douglas, 2019). Further research of these and other systems are needed for the pursuit of technological and medical advances or cultural/societal changes necessary for overall human and planetary benefit.
Microbiome Analysis
Traditionally, it has been difficult to characterize complete microbial communities, as most projects require substantial time and resources. Though, advancements in NGS and ‘omics’ technologies have begun to allow researchers to tackle microbiome analytics using a variety of approaches including metagenomics, metatranscriptomics, proteomics, metabolomics, culturomics, etc.(Integrative HMP (iHMP) Research Network Consortium, 2014, Bashiardes et al., 2016, Daliri et al., 2017, Janson and Hofmockel, 2018, Lin et al., 2019, Diakite et al., 2020, ). Each technique is selected and applied depending on the experimental setup and what questions are being addressed. For instance, does the research care about the identification of members of the microbiome, how they are interacting with the host or each other, what macromolecules are present, what genes are being expressed, what is the functional potential, etc. These different types of investigations can then be integrated together in network analyses to establish linkages and correlations within the microbiome datasets, though statistical models and analytical tools must be carefully selected to avoid false outcomes and shortcomings (Jiang et al., 2019). Due to the particular limitations of these approaches and the unavoidably large datasets produced by microbiomic studies, it is paramount to continually develop novel technologies to unearth the knowledge buried in these microbiomes.
Considering the abundance of microbiomes that no doubt play role in human health, the extent to which microorganisms sway our lives is almost impossible to foresee, and the opportunities they present for improvement to industry, agriculture, environmental and human health is potentially unlimited.
Check Your Understanding
- What is the difference between a microbiome and microbiota?
- In what ways does a microbiome influence a holobiont?
- How can an environmental microbiome indirectly impact human health?
Media Attributions
- Video 1 – What is the human microbiome? by The Conversation. Licensed under Creative Commons: By Attribution 3.0 License https://creativecommons.org/licenses/by/3.0/
- Video 2 – Human Science (Part I) – The Gut Brain Axis, Microbiome & the power of Probiotics by Infognostica. Licensed under Creative Commons: By Attribution 3.0 License https://creativecommons.org/licenses/by/3.0/
- Figure 1 – Microbiome Schematic by Berg et al., 2020 adapted by Dylan Parks. Licensed under Creative Commons: By Attribution 4.0 License http://creativecommons.org/licenses/by/4.0/
- Figure 2 – History of Microbiome Research by Berg et al., 2020. Licensed under Creative Commons: By Attribution 4.0 License http://creativecommons.org/licenses/by/4.0/
References
- Apprill, A. (2017). Marine Animal Microbiomes: Toward Understanding Host–Microbiome Interactions in a Changing Ocean. Frontiers in Marine Science, 4, 222. https://doi.org/10.3389/fmars.2017.00222
- Arif, I., Batool, M., & Schenk, P. M. (2020). Plant Microbiome Engineering: Expected Benefits for Improved Crop Growth and Resilience. Trends in Biotechnology, 38(12), 1385–1396. https://doi.org/10.1016/j.tibtech.2020.04.015
- Backhed F, Fraser CM, Ringel Y, Sanders ME, Sartor RB, Sherman PM, et al. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe. 2012;12:611–22. https://doi.org/10.1016/j.chom.2012.10.012
- Baedke, J., Fábregas-Tejeda, A., & Nieves Delgado, A. (2020). The holobiont concept before Margulis. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 334(3), 149–155. https://doi.org/10.1002/jez.b.22931
- Bashiardes S, Zilberman-Schapira G, Elinav E. Use of Metatranscriptomics in Microbiome Research. Bioinformatics and Biology Insights. January 2016. doi:10.4137/BBI.S34610
- Berg, G., Rybakova, D., Fischer, D. et al. Microbiome definition re-visited: old concepts and new challenges. Microbiome 8, 103 (2020). https://doi.org/10.1186/s40168-020-00875-0
- Busby PE, Soman C, Wagner MR, Friesen ML, Kremer J, Bennett A, et al. (2017) Research priorities for harnessing plant microbiomes in sustainable agriculture. PLoS Biol 15(3): e2001793. https://doi.org/10.1371/journal.pbio.2001793
- Clapp M, Aurora N, Herrera L, Bhatia M, Wilen E, Wakefield S. Gut Microbiota’s Effect on Mental Health: The Gut-Brain Axis. Clinics and Practice. 2017; 7(4):131-136. https://doi.org/10.4081/cp.2017.987
- Cryan, J. F., O’Riordan, K. J., Cowan, C. S. M., Sandhu, K. v, Bastiaanssen, T. F. S., Boehme, M., Codagnone, M. G., Cussotto, S., Fulling, C., Golubeva, A. v, Guzzetta, K. E., Jaggar, M., Long-Smith, C. M., Lyte, J. M., Martin, J. A., Molinero-Perez, A., Moloney, G., Morelli, E., Morillas, E., … Dinan, T. G. (2019). The Microbiota-Gut-Brain Axis. Physiological Reviews, 99(4), 1877–2013. https://doi.org/10.1152/physrev.00018.2018
- Diakite, A., Dubourg, G., Dione, N. et al. Optimization and standardization of the culturomics technique for human microbiome exploration. Sci Rep 10, 9674 (2020). https://doi.org/10.1038/s41598-020-66738-8
- Elhady, A., Adss, S., Hallmann, J., & Heuer, H. (2018). Rhizosphere Microbiomes Modulated by Pre-crops Assisted Plants in Defense Against Plant-Parasitic Nematodes. Frontiers in Microbiology, 9, 1133. https://www.frontiersin.org/article/10.3389/fmicb.2018.01133
- Daliri, E. B., Wei, S., Oh, D. H., & Lee, B. H. (2017). The human microbiome and metabolomics: Current concepts and applications. Critical reviews in food science and nutrition, 57(16), 3565–3576. https://doi.org/10.1080/10408398.2016.1220913
- Foster, J. A., Rinaman, L., & Cryan, J. F. (2017). Stress & the gut-brain axis: Regulation by the microbiome. Neurobiology of Stress, 7, 124–136. https://doi.org/https://doi.org/10.1016/j.ynstr.2017.03.001
- Garrett W. S. (2015). Cancer and the microbiota. Science (New York, N.Y.), 348(6230), 80–86. https://doi.org/10.1126/science.aaa4972
- Gilbert, J. A., Blaser, M. J., Caporaso, J. G., Jansson, J. K., Lynch, S. v, & Knight, R. (2018). Current understanding of the human microbiome. Nature Medicine, 24(4), 392–400. https://doi.org/10.1038/nm.4517
- Grice, E., Segre, J. The skin microbiome. Nat Rev Microbiol 9, 244–253 (2011). https://doi.org/10.1038/nrmicro2537
- Hannula, S., Morriën, E., de Hollander, M. et al. Shifts in rhizosphere fungal community during secondary succession following abandonment from agriculture. ISME J 11, 2294–2304 (2017). https://doi.org/10.1038/ismej.2017.90
- Hernández-Álvarez, C., García-Oliva, F., Cruz-Ortega, R., Romero, M. F., Barajas, H. R., Piñero, D., & Alcaraz, L. D. (2022). Squash root microbiome transplants and metagenomic inspection for in situ arid adaptations. Science of The Total Environment, 805, 150136. https://doi.org/https://doi.org/10.1016/j.scitotenv.2021.150136
- Hirt, H. (2020). Healthy soils for healthy plants for healthy humans. EMBO Reports, 21(8), e51069. https://doi.org/https://doi.org/10.15252/embr.202051069
- Hsiao, E. Y., McBride, S. W., Hsien, S., Sharon, G., Hyde, E. R., McCue, T., Codelli, J. A., Chow, J., Reisman, S. E., Petrosino, J. F., Patterson, P. H., & Mazmanian, S. K. (2013). Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell, 155(7), 1451–1463. https://doi.org/10.1016/j.cell.2013.11.024
- Lin, H., He, Q. Y., Shi, L., Sleeman, M., Baker, M. S., & Nice, E. C. (2019). Proteomics and the microbiome: pitfalls and potential. Expert review of proteomics, 16(6), 501–511. https://doi.org/10.1080/14789450.2018.1523724
- Human Microbiome Project / Program Initiatives. The NIH Common Fund. Retrieved 9 September 2021. https://commonfund.nih.gov/hmp/initiatives
- Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, et al. Human Microbiome Project Consortium (2012). Structure, function and diversity of the healthy human microbiome. Nature, 486(7402), 207–214. https://doi.org/10.1038/nature11234
- Integrative HMP (iHMP) Research Network Consortium (2014). The Integrative Human Microbiome Project: dynamic analysis of microbiome-host omics profiles during periods of human health and disease. Cell host & microbe, 16(3), 276–289. https://doi.org/10.1016/j.chom.2014.08.014
- Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D. Role of the normal gut microbiota. World J Gastroenterol. 2015 Aug 7;21(29):8787-803. doi: 10.3748/wjg.v21.i29.8787. PMID: 26269668; PMCID: PMC4528021. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4528021/
- Jansson, J. K., & Hofmockel, K. S. (2018). The soil microbiome—from metagenomics to metaphenomics. Current Opinion in Microbiology, 43, 162–168. https://doi.org/10.1016/j.mib.2018.01.013
- Jiang, D., Armour, C. R., Hu, C., Mei, M., Tian, C., Sharpton, T. J., & Jiang, Y. (2019). Microbiome Multi-Omics Network Analysis: Statistical Considerations, Limitations, and Opportunities. Frontiers in genetics, 10, 995. https://doi.org/10.3389/fgene.2019.00995
- Lederberg J, Mccray AT. `Ome Sweet `Omics–A genealogical treasury of words. The Scientist. 2001;15(7):8–8. https://lhncbc.nlm.nih.gov/LHC-publications/pubs/OmeSweetOmicsAGenealogicalTreasuryofWords.html
- Levkovich T, Poutahidis T, Smillie C, Varian BJ, Ibrahim YM, Lakritz JR, et al. (2013) Probiotic Bacteria Induce a ‘Glow of Health’. PLoS ONE 8(1): e53867. https://doi.org/10.1371/journal.pone.0053867
- Lloyd-Price, J., Abu-Ali, G. & Huttenhower, C. The healthy human microbiome. Genome Med 8, 51 (2016). https://doi.org/10.1186/s13073-016-0307-y
- Marchesi, J. R., & Ravel, J. (2015). The vocabulary of microbiome research: a proposal. Microbiome, 3, 31. https://doi.org/10.1186/s40168-015-0094-5
- Margulis L. Symbiosis as a source of evolutionary innovation: speciation and morphogenesis. In: Cambridge MA MLFR, editor. Symbiogenesis and Symbionticism: MIT Press; 1991. p. 1–14.
- NIH Human Microbiome Project – About the Human Microbiome. https://hmpdacc.org/ihmp/overview/. Retrieved 9 September 2021.
- O’Neill, C.A., Monteleone, G., McLaughlin, J.T. and Paus, R. (2016), The gut-skin axis in health and disease: A paradigm with therapeutic implications. BioEssays, 38: 1167-1176. https://doi.org/10.1002/bies.201600008
- Petersen, C., & Round, J. L. (2014). Defining dysbiosis and its influence on host immunity and disease. Cellular microbiology, 16(7), 1024–1033. https://doi.org/10.1111/cmi.12308
- Pratama, A. A., & van Elsas, J. D. (2018). The ‘Neglected’ Soil Virome – Potential Role and Impact. Trends in Microbiology, 26(8), 649–662. https://doi.org/https://doi.org/10.1016/j.tim.2017.12.004
- Rosenberg, E. and Zilber-Rosenberg, I. (2011), Symbiosis and development: The hologenome concept. Birth Defects Research Part C: Embryo Today: Reviews, 93: 56-66. https://doi.org/10.1002/bdrc.20196
- Saleem, M., Hu, J., & Jousset, A. (2019). More Than the Sum of Its Parts: Microbiome Biodiversity as a Driver of Plant Growth and Soil Health. Annual Review of Ecology, Evolution, and Systematics, 50(1), 145–168. https://doi.org/10.1146/annurev-ecolsys-110617-062605
- Salem I, Ramser A, Isham N and Ghannoum MA (2018) The Gut Microbiome as a Major Regulator of the Gut-Skin Axis. Front. Microbiol. 9:1459. doi: 10.3389/fmicb.2018.01459
- Sender R, Fuchs S, Milo R (2016) Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol 14(8): e1002533. https://doi.org/10.1371/journal.pbio.1002533
- Simon, JC., Marchesi, J.R., Mougel, C. et al. Host-microbiota interactions: from holobiont theory to analysis. Microbiome 7, 5 (2019). https://doi.org/10.1186/s40168-019-0619-4
- The Integrative HMP (iHMP) Research Network Consortium. The Integrative Human Microbiome Project. Nature 569, 641–648 (2019). https://doi.org/10.1038/s41586-019-1238-8
- Trompette, A., Gollwitzer, E. S., Yadava, K., Sichelstiel, A. K., Sprenger, N., Ngom-Bru, C., Blanchard, C., Junt, T., Nicod, L. P., Harris, N. L., & Marsland, B. J. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nature medicine, 20(2), 159–166. https://doi.org/10.1038/nm.3444
- Turnbaugh, P., Ley, R., Hamady, M. et al. The Human Microbiome Project. Nature 449, 804–810 (2007). https://doi.org/10.1038/nature06244
- Whipps J, Lewis K, Cooke R. Mycoparasitism and plant disease control. In: Burge M, editor. Fungi Biol Control Syst. Manchester University Press; 1988. p. 161-187.
- Zhong, W., Yian, G., Ville-Petri, F., A, K. G., Yangchun, X., Qirong, S., & Alexandre, J. (2021). Initial soil microbiome composition and functioning predetermine future plant health. Science Advances, 5(9), eaaw0759. https://doi.org/10.1126/sciadv.aaw0759

